Association Between Gut Microbiota and Helicobacter pylori-Related Gastric Lesions in a High-Risk Population of Gastric Cancer
- 1Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education), Department of Cancer Epidemiology, Peking University Cancer Hospital and Institute, Beijing, China
- 2Institute of Medical Microbiology, Immunology and Hygiene, Technische Universität München, Munich, Germany
- 3German Center for Infection Research, Partner Site Munich, Munich, Germany
- 4Institute of Pathology, Klinikum Bayreuth, Bayreuth, Germany
- 5II. Medizinische Klinik, Klinikum Rechts der Isar, Technische Universität München, Munich, Germany
- 6Department of Internal Medicine, 1st Faculty of Medicine, Charles University, Military University Hospital, Prague, Czechia
- 7Linqu Public Health Bureau, Linqu, Shandong, China
- 8International Digestive Cancer Alliance, Munich, Germany
Eradication of Helicobacter pylori has been found to be effective for gastric cancer prevention, but uncertainties remain about the possible adverse consequences such as the potential microbial dysbiosis. In our study, we investigated the association between gut microbiota and H. pylori-related gastric lesions in 47 subjects by deep sequencing of microbial 16S ribosomal RNA (rRNA) gene in fecal samples. The dominant phyla in fecal samples were Bacteroidetes, Firmicutes, and Proteobacteria with average relative abundances of 54.77, 31.37 and 12.91%, respectively. Microbial diversity analysis showed that observed species and Shannon index were increased in subjects with past or current H. pylori infection compared with negative subjects. As for the differential bacteria, the average relative abundance of Bacteroidetes was found to significantly decrease from H. pylori negative (66.16%) to past infection group (33.01%, p = 0.007), as well as from normal (76.49%) to gastritis (56.04%) and metaplasia subjects (46.83%, p = 0.027). For Firmicutes and Proteobacteria, the average relative abundances showed elevated trends in the past H. pylori infection group (47.11, 20.53%) compared to negative group (23.44, 9.05%, p = 0.068 and 0.246, respectively), and similar increased trends were also found from normal (18.23, 5.05%) to gastritis (35.31, 7.23%, p = 0.016 and 0.294, respectively) or metaplasia subjects (32.33, 20.07%, both p < 0.05). These findings suggest that the alterations of fecal microbiota, especially the dominant phyla of Bacteroidetes, Firmicutes and Proteobacteria, may be involved in the process of H. pylori-related gastric lesion progression and provide hints for future evaluation of microbial changes after H. pylori eradication.
Introduction
Gastric cancer (GC) is a major health problem in China, accounting for over 40% of the new GC cases annually worldwide (Torre et al., 2015). In the multistep gastric carcinogenesis process, Helicobacter pylori plays a crucial role by inducing inflammation and degradation of the gastric epithelium from early stage (Correa, 1992). The factors affecting the outcome of H. pylori infection are diverse and include bacterial virulence, host genetic factors and host microbiota (Lofgren et al., 2011).
The human gastrointestinal microbiota is a complex and dynamic ecosystem regarded as a metabolically active “organ.” Thus, cosstalk between microbes and immune cells regulates human gastrointestinal homeostasis (Mortha et al., 2014). The balance of commensal microbiota in the gastrointestinal tract may play important roles for regulation of host mucosal immune response, energy metabolism, elimination of pathogens and cancer development (Garrett, 2015; Rooks and Garrett, 2016). Many previous studies suggested that gastric microbiota can be affected by H. pylori infection (Maldonado-Contreras et al., 2011; Jo et al., 2016; Schulz et al., 2018). Dysbiosis of gastric microbiota and some specific bacteria were found to be associated with GC or precancerous lesions (Eun et al., 2014; Coker et al., 2018; Ferreira et al., 2018). In addition, a distinct shift of microbiota in the distal intestinal tract was reported following hypochlorhydria and hypergastrinemia by H. pylori infection in Mongolian gerbils (Heimesaat et al., 2014). However, little is known about the association between intestinal microbiota and H. pylori infection or GC and precancerous lesions in human beings. Since it is well appreciated that microbiota composition shape immune responses at a local and systemic level, and also that GC development is influenced by inflammatory signaling, it is tempting to speculate that H. pylori associated alterations of the gut microbiota may in turn influence GC development.
H. pylori eradication was found to be effective in preventing precancerous gastric lesions and GC in previous intervention trials (Wong et al., 2004; You et al., 2006). However, uncertainties remain about the possible adverse consequences of anti-H. pylori treatment. Several studies suggested that adverse health effects, including obesity (Ley et al., 2006), asthma (Couzin-Frankel, 2010), or other immunological conditions (Sekirov et al., 2010) may be associated with the perturbation of gut microbiome after H. pylori eradication. Thus, a deep understanding of the association between microbial dysbiosis and H. pylori-related precancerous gastric lesions is urgently needed for optimizing the efficacy of GC prevention strategies.
In the present study, we analyzed the gut microbiota of subjects presenting gastric lesions associated with or independent of H. pylori infection by deep sequencing of microbial 16S ribosomal RNA (rRNA) gene in feces. Our results provide baseline associations between gut microbiota and H. pylori-related gastric lesions, which will be important for further evaluation of eradication treatment and gastric carcinogenesis.
Materials and Methods
Study Subjects and Sample Collection
In November 2014, an endoscopy study was conducted in Linqu County, China. A total of 218 asymptomatic individuals (aged 40–69 years) from two villages of Yishan Township were selected for 13C-urea breath test (13C-UBT) and standardized upper endoscopy examination. The exclusion criteria were as follows: contraindication for biopsy sampling, risk of conscious sedation due to severe comorbidities, and inability to provide an informed consent due to psychiatric illness. In the final week of the endoscopic examinations, participants were invited to provide stool samples. A total of 47 participants agreed and provided samples, which were collected and frozen immediately at −80°C until DNA extraction. Besides, each participant completed a physical examination and a questionnaire on age, gender, cigarette smoking, alcohol consumption, and antibiotics treatment history under the supervision of a well-trained interviewer before the endoscopic examination. This study was approved by the Institutional Review Boards of Peking University Cancer Hospital and Institute and written informed consent was obtained from all of the participants.
Upper Endoscopic Examination and Histopathology
Upper endoscopic examinations were conducted by five international experienced gastroenterologists using video endoscopes (Olympus). The gastric mucosa was examined, and five or more biopsies were obtained from antrum, corpus, angulus, cardia, esophagus and any suspicious lesions. All gastric mucosa specimens were reviewed blindly by two pathologists (1 Chinese and 1 German) according to the Updated Sydney System (Dixon et al., 1996). Each biopsy was scored using a visual analog scale (0 = absent, 1 = mild, 2 = moderate, 3 = severe) for H. pylori infection, gastritis, activity of inflammation and intestinal metaplasia (Dixon et al., 1996). Each subject was assigned a global diagnosis for each variable based on the most severe diagnosis reported from all the biopsies.
Determination of H. pylori Infection Status
H. pylori infection status was determined according to 13C-UBT and histological diagnosis. 13C-UBT was performed as reported previously (Pan et al., 2016). Subjects with concentration of 13CO2 that exceeded the baseline >3.8 parts per 1000 (>0.38%) after 30 min were regarded as positive. Current H. pylori infection was defined when bacteria were histologically detected in any one of the biopsies or it was positively indicated by 13C-UBT. Non-current infection subjects were defined when both bacterial and 13C-UBT detection indicated negative results.
From the non-current infection subjects, past infection subjects were identified by record on previous H. pylori eradication therapy or histological diagnosis of non-active gastritis with basal lymphoid aggregates and reactive changes of the surface epithelium in antrum and corpus, indicating a prior H. pylori infection. Non-current infection subjects without eradication treatment history and specific histological diagnosis were considered negative H. pylori infection subjects.
Nucleic Acid Extraction From Stool Samples
Fecal DNA was extracted using the Qiagen Fast DNA Stool Mini Kit (Qiagen, MD, USA) according to the protocol provided by the manufacturer. Briefly, 180–220 mg sample was re-suspended in InhibitEX Buffer to adsorb DNA-damaging substances and PCR inhibitors. After thoroughly homogenizing, the suspension was heated to 95°C for 5 min to maximize the lysis of bacteria. The quantity and quality of total DNA were verified by NanoDrop-2000 spectrophotometer (Thermo Scientific) and 1% standard agarose gel electrophoresis.
16S rRNA Gene Sequencing
For analysis of microbial composition, the hypervariable region V4 of microbial 16S rRNA gene was amplified using Phusion® High-Fidelity PCR Master Mix (New England Biolabs) and the universal primers (515F, 5′-GTGCCAGCMGCCGCGGTAA-3′; 806R, 5′-GGACTACHVGGGTWTCTAAT-3′) (Baker et al., 2003; Huws et al., 2007). Thirty cycles were performed. Then PCR products were analyzed by 2% agarose gel electrophoresis, pooled in equidensity ratios, and purified with GeneJET Gel Extraction Kit (Thermo Scientific). Sequencing libraries were generated using NEB Next® Ultra® DNA Library Prep Kit for Illumina (NEB, USA) following manufacturer's recommendations. Library quality was assessed using a Qubit@ 2.0 Fluorometer (Thermo Scientific) and an Agilent Bioanalyzer 2100 system. The library was sequenced on an Illumina MiSeq platform (Novogene Bioinformatics Technology Co., Ltd. Beijing, China) to produce 250-bp paired-end reads.
Data and Statistical Analysis
Paired-end reads were merged by FLASH (V1.2.7) (Magoc and Salzberg, 2011). The raw sequences were filtered to obtain high-quality sequences according to QIIME (Version 1.7.0) (Bokulich et al., 2013). UCHIME algorithm (Edgar et al., 2011) was used to detect and remove chimeras. Sequences with ≥97% similarity were assigned to the same operational taxonomic units (OTUs) by UPARSE software package (UPARSE, v7.0.1001) (Edgar, 2013). A representative sequence for each OTU was picked and annotated using the Mothur method based on the SSUrRNA database of SILVA (Quast et al., 2013) (setting a threshold of 0.8–1) to obtain classification levels (phylum, class, order, family, genus and species). The abundance information of OTUs was normalized with a standard sequence number corresponding to the sample with the fewest sequences, based on which subsequent analyses were performed.
The microbial alpha diversity of fecal samples was illustrated by observed species and Shannon index, which were calculated using QIIME (Version 1.7.0). Rarefaction curves were displayed with R software (Version 2.15.3). Comparisons of microbial communities were visualized by heat-map and Principal Co-ordinates Analysis (PCoA) based on weighted unifrac distances between clinical parameters and gut microbiota, and the significance of differences was evaluated by the analysis of similarity (ANOSIM) depending on the Bray-Curtis dissimilarity distance matrices. Taxa with relative abundance more than 0.1% in at least one group (without special emphasis) were included in identification of differential taxa. Welch's t-test was utilized to identify differences in the average relative abundances of individual taxonomy between two groups (Parks et al., 2014).
Continuous variables, such as age, body mass index (BMI) and fasting plasma glucose, were reported as mean ± standard deviation, while observed species and Shannon index were reported as median (interquartile range, IQR). Categorical variables (gender, smoking, drinking, antibiotics treatment history and gastric lesions) were described as frequencies and proportions. Comparisons of continuous parameters were performed by Mann-Whitney test between two groups or Kruskal-Wallis test among three groups. Fisher's exact test was conducted for comparisons between categorical variables. Statistical analysis was carried out using SPSS 13.0 (SPSS, USA), and p < 0.05 were considered statistically significant.
Results
From 47 subjects who provided fecal samples, 24 subjects were identified as having current H. pylori infection and 23 subjects as non-current infection, of which 15 were H. pylori negative and 8 showed signs of past infection (Table 1). The frequency of metaplasia was marginally higher in the current H. pylori infection and in the past infection groups than in the negative group (62.50, 50.00, and 20.00%, p = 0.065). No significant differences were found among the three groups in age, gender, BMI, smoking, drinking, fasting plasma glucose and antibiotics use (all p > 0.05).
Fecal Microbiota Composition
A total of 2,827,158 paired-end reads were generated from 47 fecal samples including 2,557,652 quality-filtered reads with an average of 54,418 ± 5,665 (standard deviation) reads per sample. After aligning to the SILVA database and removing non-bacterial sequences, 2,518,760 (98.48%) of the quality-filtered reads were finally retained for subsequent analysis with an average of 53,591 ± 5,625 reads per sample, which generated 817 OTUs at 97% similarity level. The rarefaction curves showed a reasonable sequencing depth (Figure S1).
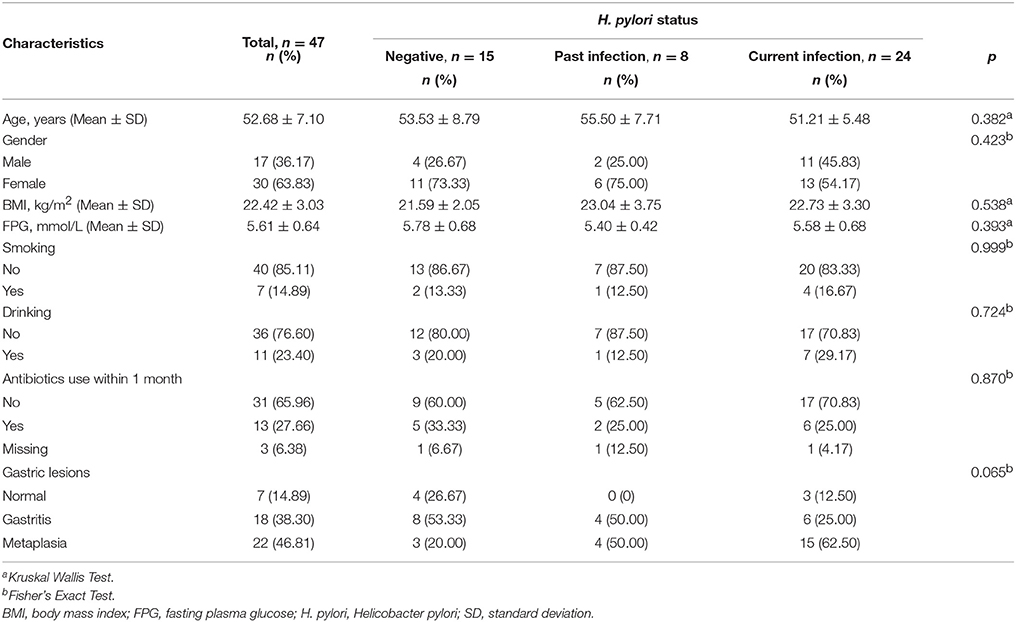
Phyla and genera of fecal microbiota detected in the 47 subjects are summarized in Figure 1. The most abundant phyla were Bacteroidetes, Firmicutes, and Proteobacteria with average relative abundances of 54.77%, 31.37% and 12.91%, respectively. The three dominant phyla accounted for 99.05% of all fecal bacteria (Figure 1A). At the genus level, fecal microbiota was dominated by Bacteroides, Prevotella_9, Escherichia-Shigella and Ruminococcus_2 with average relative abundances of 33.53, 12.55, 5.07, and 4.34%, respectively (Figure 1B). Only one sequence of Archaea was found in two of the fecal samples with relative abundances <0.10% (Table S1).
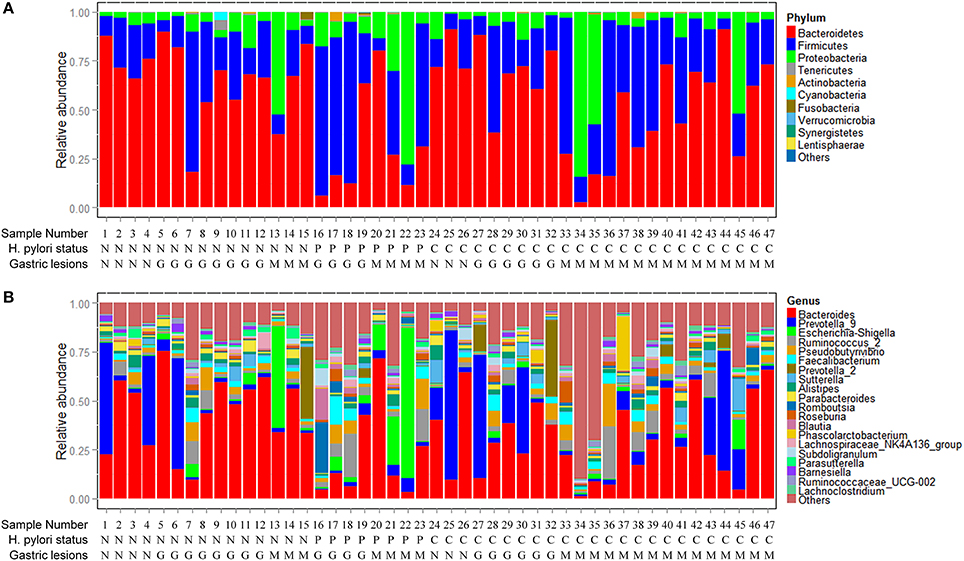
Figure 1. Composition of fecal microbiota. (A) Relative abundance distribution of major phyla (top 10) in 47 fecal samples. Phyla were sorted by decreasing order of average relative abundance. H. pylori status and gastric lesions observed for each subject are listed below. (B) Relative abundance distribution of major genera (top 20) in 47 fecal samples. Genera were sorted by decreasing order of average relative abundance as in (A). H. pylori status: N, H. pylori negative; P, past H. pylori infection; C, current H. pylori infection. Gastric lesions: N, normal; G, gastritis; M, metaplasia.
Association Between H. pylori Infection and Fecal Microbiota
To explore the association between H. pylori infection and fecal microbiota, we firstly compared the microbial alpha diversity, community structure, and differential taxa between current and non-current H. pylori infection groups. For alpha diversity analysis, we used observed species to evaluate microbial richness and Shannon index to evaluate evenness. No statistical differences were found in medians (IQR) of observed species [316.00 (285.75–351.50) vs. 324.00 (289.00–351.00), p = 0.831] and Shannon index [4.83 (3.73–5.36) vs. 4.79 (3.99–5.44), p = 0.983] between non-current H. pylori infection and current infection group (Figures S2A,B). Likewise, no significant difference in fecal bacterial community structure was found between the current and non-current H. pylori infection groups (R = 0.007, p = 0.300, Table S3; Figure S2C).
When we compared the differential taxa between the two groups, no significant differences were found in major phyla. At the genus level, average relative abundances were found significantly decreased for Acidovorax and Rhodococcus (p = 0.016 and 0.017, respectively), and increased for Gemella and Erysipelotrichaceae_UCG_004 (p = 0.002 and 0.020, respectively) in the current infection group when compared to the non-current infection group (Figure S2D). However, the average relative abundances of these differential genera were only <0.1% of the fecal microbiota.
Non-current H. pylori infection subjects were divided into negative (n = 15) and past infection (n = 8) groups according to antibiotics treatment history and histological assessment to further compare the fecal microbiota among the different groups. Observed species [median (IQR) 317.00 (289.00–328.00)] and Shannon index [4.74 (3.99–5.27)] were increased from negative, to past infection [351.00 (296.00–362.00), 5.27 (3.46–5.59)] or current infection group [316.00 (285.75–351.50), 4.83 (3.73–5.36)], but the differences were not statistically significant (p = 0.317 for observed species and 0.696 for Shannon index) (Table S2, Figures 2A,B).
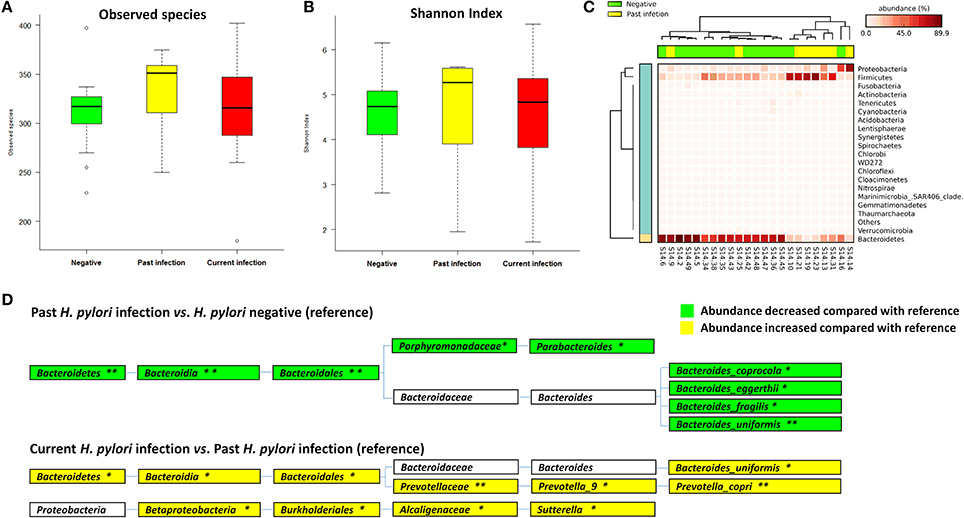
Figure 2. Associations between Helicobacter pylori infection and fecal microbiota. (A) Boxplot of observed species in different H. pylori status groups. The boxes indicate interquartile ranges (IQRs) and the median (blank line).Whiskers extend to the most extreme points within 1.5-fold IQR. Outliers are plotted individually (°). (B) Shannon index in different H. pylori status groups. (C) Heat-map of relative abundance distributions of the phyla in past H. pylori infection (n = 8) and negative groups (n = 15). (D) Cladogram of differently distributed taxa between groups. The differential taxa are illustrated between past H. pylori infection and negative groups, or current H. pylori infection and past infection groups as *p < 0.05 and **p < 0.01, respectively.
A significant difference in fecal microbial community structure was found by ANOSIM between past H. pylori infection and negative groups (R = 0.316, p = 0.004, Table S3, Figure S3A), while no statistical difference was found between the current H. pylori infection and negative groups (R = −0.038, p = 0.763, Table S3, Figure S3A). The heat-map based on relative abundances of all phyla also revealed that past H. pylori infection subjects were distinct from subjects of the negative group (Figure 2C).
We further studied differential bacteria among the three different H. pylori infection groups (Figure 2D). The average relative abundance of Bacteroidetes (including the genera of Parabacteroides and some species belonging to Bacteroides) was significantly decreased from 66.16% in the H. pylori negative group to 31.01% in the past infection group, p = 0.007. The average relative abundance of Bacteroidetes was also found increased from 31.01% in the past H. pylori infection group to 55.57% in the current infection group, p = 0.043. Especially Bacteroides uniformis species showed significantly decreased relative abundance in the past H. pylori infection group (1.50%), when compared to the current infection (3.74%, p = 0.022) and the negative groups (5.37%, p = 0.002), respectively.
Association Between Gastric Lesions and Fecal Microbiota
The global gastric pathological diagnosis of the 47 subjects included in the study ranged from normal to gastritis and metaplasia. This provided the opportunity to investigate the relationship between gastric lesions and fecal microbiota. Microbial alpha diversity analysis revealed no significant differences in observed species and Shannon index medians among the normal [301.00 (284.00–320.00), 4.68 (3.85–4.84)], gastritis [327.00 (309.75–351.50), 5.03 (4.26–5.61)] and metaplasia groups [320.00 (281.00–358.00), 4.87 (3.42–5.26), both p > 0.05] (Table S2). When compared to the normal group as reference, observed species and Shannon index were increased in gastritis and metaplasia groups, but the differences were not statistically significant (p = 0.397 in gastritis, p = 0.759 in metaplasia for observed species, and p = 0.258, p = 0.983 for Shannon index) (Figures 3A,B).
Figure 3. Associations of gastric lesions with fecal microbiota. (A) Boxplot of observed species in different gastric lesion groups. (B) Boxplot of Shannon index in different gastric lesion groups. (C) Heat-map demonstrating the relative abundance of different phyla in gastritis (n = 18) and normal (n = 7) samples. (D) Heat-map showing the relative abundance distributions of different phyla in metaplasia (n = 22) and normal samples.
No significant difference in fecal microbial community structure was found among the different gastric lesions groups by ANOSIM (R = −0.029, p = 0.704, Table S3, Figure S3B), while the heat-map based on the relative abundance of all phyla still showed differences between normal subjects and cases with gastritis or metaplasia (Figures 3C,D).
The differential bacteria observed in the different gastric lesions groups are shown in Figure 4. Compared to normal subjects, the differential phyla of fecal microbiota in gastritis cases were Bacteroidetes, with decreased average relative abundance from 76.49% (normal) to 56.04% (gastritis), p = 0.009, and Firmicutes with increased average relative abundance from 18.23 to 35.31%, p = 0.016. When we compared metaplasia cases with normal subjects, similar significant differences in average relative abundances were also found for Bacteroidetes (metaplasia: 46.83% vs. normal: 76.49%, p < 0.001) and Firmicutes (metaplasia: 32.33% vs. normal: 18.23%, p = 0.016). In addition, Proteobacteria showed significantly increased average relative abundance from normal (5.05%) to metaplasia subjects (20.07%, p = 0.016) and from gastritis (7.23%) to metaplasia (20.07%, p = 0.034). Finally, the average relative abundance of Enterobacteriaceae, a family of Proteobacteria, was significantly increased with the severity of the gastric lesions analyzed (normal: 0.60%, gastritis: 2.95%, metaplasia: 16.14%, p = 0.033).

Figure 4. Cladogram of differentially distributed taxa among different gastric lesion groups. The differential taxa are illustrated between gastritis and normal, metaplasia and normal, or metaplasia and gastritis groups as *p < 0.05, **p < 0.01, and ***p < 0.001, respectively.
Association Between Activity of Gastritis and Fecal Microbiota
Since neutrophil activity is a very sensitive indicator for current H. pylori-related gastritis, we further analyzed the association between activity of gastritis and fecal microbiota. Of the 18 subjects diagnosed with gastritis, 12 presented non-active gastritis and 6 showed active gastritis. Analyses of observed species and Shannon index showed that the microbial richness and evenness in fecal samples were slightly increased in non-active gastritis subjects [332.50 (312.25–352.50), 5.03 (4.72–5.61)] compared to normal subjects [301.00 (284.00–320.00), 4.68 (3.85–4.84)], although p values were 0.098 and 0.184, respectively (Table S2, Figures S4A,B). No significant differences were found between subjects presenting active gastritis [316.00 (240.00–346.25), 4.81 (3.55–5.46)] and normal subjects [301.00 (284.00–320.00), 4.68 (3.85–4.84), p = 0.803, and 0.739, respectively].
No significant differences among normal, non-active and active gastritis groups (R = 0.063, p = 0.194) were detected when comparing the fecal microbiota community structure by ANOSIM (Table S3) and heat-map analysis (Figures S4C,D). In contrast, for differential taxa selection, the average relative abundance of Ruminococcaceae_NK4A214_group was found decreased in active gastritis (0.06%) compared to non-active gastritis subjects (0.16%, p = 0.030) (Figure S4E).
Identification of Relevant Fecal Microbial Alterations in H. pylori Infection and Gastric Lesions
To further identify associations between fecal microbiota and H. pylori-related gastric lesions, we finally analyzed relevant microbial alterations in relation to H. pylori infection and progression of gastric lesions. In the three dominant phyla (Bacteroidetes, Firmicutes, and Proteobacteria) of fecal microbiota (Figure 5A), the average relative abundance of Bacteroidetes decreased from normal (76.49%) to gastritis (56.04%) and metaplasia (46.83%, p = 0.027), as well as from H. pylori negative (66.16%) to past infection (33.01%, p = 0.007). For Firmicutes and Proteobacteria, the average relative abundances were increased from normal (18.23, 5.05%) to gastritis (35.31, 7.23%, p = 0.016 and 0.294) or metaplasia (32.33, 20.07%, both p < 0.05), respectively. Similarly, the average relative abundances of these two phyla were found elevated in past H. pylori infection (47.11, 20.53%) compared to the negative group (23.44, 9.05%), although the p values (0.068 and 0.246) showed no significant differences.
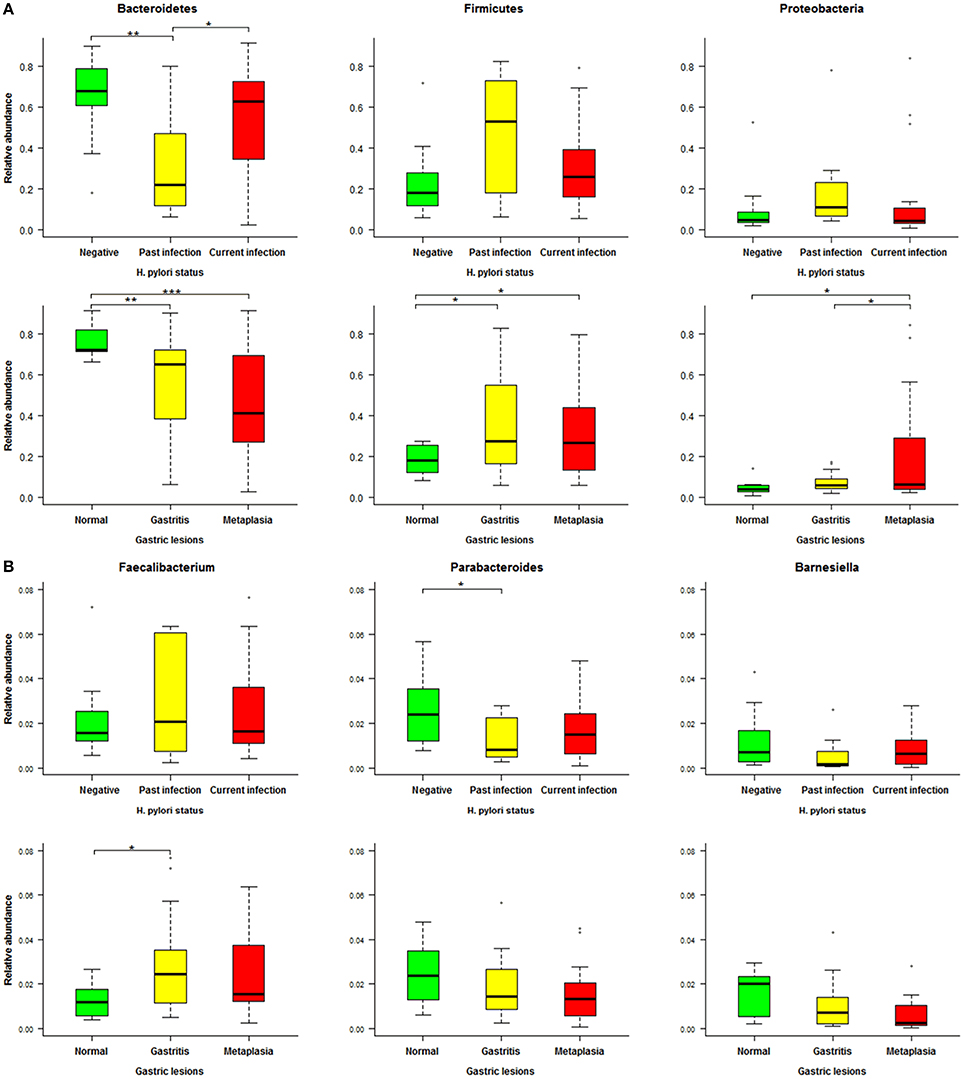
Figure 5. Relative abundance distributions of specific phyla and genera in different Helicobacter pylori infection status and gastric lesion groups. (A) Boxplot of the main phyla in different H. pylori infection status and gastric lesion groups. (B) Boxplot of specific genera in different H. pylori infection status and gastric lesion groups. Significant differences between groups are indicated as ***p < 0.001, **p < 0.01, and *p < 0.05, respectively.
At the genus level, a total of 19 genera with average relative abundances of more than 0.01% were analyzed in relation to H. pylori infection status and gastric lesions. Three genera (Parabacteroides, Barnesiella, and Faecalibacterium) were found to have similar alterations in H. pylori infection and gastric lesion groups, although most of the p values were higher than 0.05 among groups with different H. pylori infection status or gastric lesions (Figure 5B). The average relative abundances of Parabacteroides and Barnesiella (belonging to the phylum of Bacteroidetes) decreased with the severity of gastric lesions (normal: 2.48 and 1.56%, gastritis: 1.97 and 1.01%, metaplasia: 1.56 and 0.61%, respectively), as well as H. pylori infection status (negative: 2.44 and 1.15%, past infection: 1.27 and 0.58%, respectively). In addition, Faecalibacterium (belonging to the phylum of Firmicutes) showed a trend to higher average relative abundance both in the gastritis (3.45%) and the past H. pylori infection groups (4.04%) compared to their reference (normal: 1.27%, negative: 2.07%), respectively.
Discussion
Deep understanding of the association between gut microbiota and H. pylori-related precancerous gastric lesions is very important for evaluation of overall benefits and adverse effects of eradication treatment and for the optimization of GC prevention strategies. In the present study, we profiled fecal microbiota in subjects with different H. pylori infection status. Diversity analysis showed slightly increased microbial richness and evenness in subjects presenting signs of past H. pylori infection or gastritis compared to controls. In addition, the relative abundances of the dominant phyla Bacteroidetes, Firmicutes, and Proteobacteria in fecal microbiota were similarly altered in subjects presenting with H. pylori infection and gastric precancerous lesions. These observations suggest that fecal microbiota may be associated with the progression of H. pylori-related gastric lesions.
The human gut is inhabited by a huge number of microorganisms, the composition of which was considered critical for maintenance of gastrointestinal homeostasis. The gut microbiota is quite stable within an individual, while it varies extremely inter-individually. Many factors were reported to influence the gut microbiota including antibiotics treatment and diet (Francino, 2015; Bajaj et al., 2018). When the microbial balance or “symbiosis” turns to “dysbiosis” by the influence of various factors, the normal cohabitants of the gut may transform into “pathobionts” and trigger carcinogenesis by inducing chronic inflammation and the activation of immune mechanisms (Tözün and Vardareli, 2016). Several gastric and intestinal microbes have been recently shown as procarcinogens in GC and colorectal cancer (Jo et al., 2016; Wang et al., 2016; Flemer et al., 2017; Coker et al., 2018; Ferreira et al., 2018), or probiotics enhancing immunotherapy response of cancer patients (Gopalakrishnan et al., 2018), while little has been reported about microbiota composition in precancerous lesions. In the present study we describe the composition of fecal microbiota in a high-risk population showing different precancerous gastric lesions and H. pylori infection status. We found that the dominant phyla present in feces were Bacteroidetes, Firmicutes, and Proteobacteria, accounting for 99.05% of fecal microbiota, which is consistent with previous reports (Tözün and Vardareli, 2016). Although Helicobacter species was reported to dominate gastric microbiota in actively infected subjects, we only found few Helicobacter specific sequences in fecal samples of H. pylori positive subjects, which may be due to the different environment of the intestinal tract, in which H. pylori does not survive.
H. pylori colonizes the gastric epithelium and may influence GC progression through the interaction with gastric microbiota, which was supported by the observation of enhanced gastric carcinogenesis in H. pylori-infected germfree INS-GAS mice colonized only with three commensal bacteria (ASF356 Clostridium, ASF361 Lactobacillus murinus and ASF519 Bacterioides) (Lertpiriyapong et al., 2014). Many human studies showed that gastric microbiota can be altered by H. pylori infection (Schulz et al., 2018), with relatively lower diversity and differential abundances of Proteobacteria, Firmicutes, and Actinobacteria (Bik et al., 2006; Maldonado-Contreras et al., 2011; Li et al., 2017). Although studies on associations between H. pylori infection and intestinal microbiota were sparse, some bacteria were reported to be related with H. pylori, including Bacteroides, Prevotella spp., Clostridium histolyticum, and Lactobacilli in animal models and human studies (Bühling et al., 2001; Myllyluoma et al., 2007; Heimesaat et al., 2014). In our population-based study, only a minority of genera (Acidovorax, Rhodococcus, Gemella, and Erysipelotrichaceae_UCG-004) in feces was found associated with current H. pylori infection. No significant differences in fecal microbial diversity and structure were found between subjects showing current or non-current H. pylori infection groups. These observations are in line with the results of a previous small sample size study showing no remarkable alterations of the fecal microbiota composition and structure after H. pylori eradication (Yap et al., 2016). However, our present pilot baseline study still needs validation in a subsequent larger sample size prospective trial.
In the non-current infection subjects identified by 13C-UBT and histologic assessment, we further diagnosed past H. pylori infection from negative cases by evaluating eradication treatment history or the presence of non-active gastritis with basal lymphoid aggregates and reactive changes on the surface epithelium, which may implicate prior H. pylori infection. When we compared the three H. pylori infection groups, the distributions of possible confounding factors (including age, gender, smoking, drinking, fasting plasma glucose, BMI and recent antibiotics use) showed no significant differences. Especially, the recent antibiotics use (within 1 month) was not significantly different among negative (33%), past infection (25%), and current infection groups (25%). Consistent with a mouse model study (Lofgren et al., 2011), we found a trend to relatively increased microbial diversity and decreased relative abundance of Bacteroidetes in past H. pylori infection group compared to negative and current infection cases, respectively. The similar decrease of Bacteroidetes 12 months after H. pylori eradication in a Malaysian study (Yap et al., 2016) further suggests that long-term observation should be considered to establish associations between H. pylori infection and changes in intestinal microbiota.
So far, there are few studies about the associations between gastric microbiota and precancerous gastric lesions. A metagenomic analysis found that gastric microbial diversity was gradually decreased from gastritis to intestinal metaplasia and GC (Aviles-Jimenez et al., 2014), while other studies showed that in H. pylori positive gastric mucosa microbial diversity was higher in GC than in gastritis and intestinal metaplasia (Eun et al., 2014; Wang et al., 2016). Recently, some specific bacteria including Peptostreptococcus stomatis, Slackia exigua, Parvimonas micra, Streptococcus anginosus, and Dialister pneumosintes were found to be associated with gastric carcinogenesis (Coker et al., 2018). However, no studies were yet conducted to investigate possible alterations in intestinal microbiota in relation to gastric lesions. Our study shows a slight increased trend of microbial diversity in fecal samples of subjects presenting gastritis and metaplasia when compared to individuals showing no pathological changes in the gastric mucosa. This may be related to the fact that more H. pylori affected subjects (current and past infection) were included in the gastric lesion groups than in the normal mucosa group. The significantly different taxa observed between different lesion groups suggest a possible role of intestinal microbiota in the progression of precancerous gastric lesions. Altered microbiota observed include the dominant phyla of Bacteroidetes and Firmicutes, as well as relevant gut bacteria such as Enterobacteriaceae and Faecalibacterium previously reported to be related to inflammatory diseases such as primary biliary cholangitis (Tang et al., 2018). Of note, the frequencies of previous antibiotics treatment in our study were relatively higher in gastritis (77.8%) and metaplasia groups (68.2%) than in normal (28.6%), implicating that our results still require further validation with a larger sample size.
The mechanisms by which gut microbiota contribute to carcinogenesis are still not clear. Dysbiosis of microbiota can affect the host immune system, inducing inflammation and thereby the development of cancer due to the tight interplay between bacteria and epithelial cells. Innate bacteria-sensing receptors such as Toll-like receptors (TLRs) and NOD-like receptors (Rakoff-Nahoum and Medzhitov, 2009) may bridge this interplay, which eventually can promote carcinogenesis in a chronic process. These mechanisms may also be involved in the associations between fecal microbiota and H. pylori infection and gastric lesions observed in our population. The activity of gastritis is well known for its close relationship with H. pylori infection. This close relationship is further confirmed by the similar alterations that we observed in the fecal microbiota from non-active gastritis and past infection subjects. In addition, the same alteration tendencies detected for major phyla or genera, such as decreased abundance of Bacteroidetes and increased abundances of Firmicutes or Proteobacteria, with the severity of gastric lesion and H. pylori infection status (especially past infection status), further suggest that alterations in intestinal microbiota may be involved in the progression of H. pylori-related precancerous gastric lesions and carcinogenesis.
A major strength of our study lies in the detailed information on H. pylori infection status and the availability of tissue samples with a broad spectrum of gastric lesions from an ongoing intervention trial for H. pylori eradication. Exploration of the associations between fecal microbiota and H. pylori infection and gastric lesions builds a solid basis for our future study on the changes in microbiota after H. pylori eradication. A limitation of our study is that we only had fecal samples from 47 subjects after endoscopy examination. Therefore, the small sample size limited the feasibility to adjust possible confounding factors or correct multiple comparisons for the identification of differential taxa among groups with different H. pylori infection status or gastric lesions. However, possible factors influencing the intestinal microbiota were acquired in detail at baseline, including BMI, fasting plasma glucose and antibiotics use, which were equally distributed among different groups. Confirmation with a larger sample size including participants of the intervention trial as well as subjects showing precancerous lesions or GC is still needed. In addition, the composition of gastric microbiota and its association with H. pylori-induced gastric carcinogenesis will be further studied.
In conclusion, our study profiled the composition of fecal microbiota and assessed its association with H. pylori infection and gastric lesion status. The differential bacteria identified, including Bacteroidetes, Firmicutes and Proteobacteria, showed similar alterations associated with H. pylori infection and severity of the gastric lesions. Our results provide novel insights important for future studies on H. pylori-related carcinogenesis and alterations in the gut microbiota after H. pylori eradication.
Data Availability Statement
The raw data of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
K-FP and W-CY contributed design of the study. MB and SS conducted upper endoscopy examination. LZ, J-LM, W-DL, and Z-XL contributed subject's recruitment and samples' collection. MV, KU, MQ, RS, and MC completed histological diagnosis. YZ, J-JG, and TZ carried out experiments. J-JG analyzed experiments' results. YZ and J-JG wrote the draft of the manuscript. MG, RM-L, W-QL and K-FP revised the manuscript. All authors read and approved the submitted version.
Funding
National Natural Science Foundation of China (81572811), National Basic Research Program of China (973 Program: 2010CB529303), National Key Technology Research and Development Program (2015BA13B07), German Center for Infection Research to MG. German Federal Ministry of Education and Research (BMBF) [German Research Presence in Asia: PYLOTUM, 01DO17022 (www.internationales-buero.de/media/content/PYLOTUM.pdf)], Beijing Municipal Administration of Hospitals' Ascent Plan (DFL20181102).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank all the participants who participated in this study and donated samples.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2018.00202/full#supplementary-material
Abbreviations
13C-UBT, 13C-urea breath test; ANOSIM, analysis of similarity; BMI, body mass index; GC, gastric cancer; H. pylori, Helicobacter pylori; IQR, interquartile range; OTUs, operational taxonomic units; rRNA, ribosomal RNA; SD, standard deviation; vs., versus.
References
Aviles-Jimenez, F., Vazquez-Jimenez, F., Medrano-Guzman, R., Mantilla, A., and Torres, J. (2014). Stomach microbiota composition varies between patients with non-atrophic gastritis and patients with intestinal type of gastric cancer. Sci. Rep. 4:4202. doi: 10.1038/srep04202
Bajaj, J. S., Idilman, R., Mabudian, L., Hood, M., Fagan, A., Turan, D., et al. (2018). Diet affects gut microbiota and modulates hospitalization risk differentially in an international cirrhosis cohort. Hepatology. doi: 10.1002/hep.29791. [Epub ahead of print].
Baker, G. C., Smith, J. J., and Cowan, D. A. (2003). Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods 55, 541–555. doi: 10.1016/j.mimet.2003.08.009
Bik, E. M., Eckburg, P. B., Gill, S. R., Nelson, K. E., Purdom, E. A., Francois, F., et al. (2006). Molecular analysis of the bacterial microbiota in the human stomacH. Proc. Natl. Acad. Sci. U.S.A. 103, 732–737. doi: 10.1073/pnas.0506655103
Bokulich, N. A., Subramanian, S., Faith, J. J., Gevers, D., Gordon, J. I., Knight, R., et al. (2013). Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 10, 57–59. doi: 10.1038/nmeth.2276
Bühling, A., Radun, D., Müller, W. A., and Malfertheiner, P. (2001). Influence of anti-Helicobacter triple-therapy with metronidazole, omeprazole and clarithromycin on intestinal microflora. Aliment. Pharmacol. Ther. 15, 1445–1452. doi: 10.1046/j.1365-2036.2001.01033.x
Coker, O. O., Dai, Z., Nie, Y., Zhao, G., Cao, L., Nakatsu, G., et al. (2018). Mucosal microbiome dysbiosis in gastric carcinogenesis. Gut 67, 1024–1032. doi: 10.1136/gutjnl-2017-314281
Correa, P. (1992). Human gastric carcinogenesis: a multistep and multifactorial process–First American Cancer Society Award Lecture on Cancer Epidemiology and Prevention. Cancer Res. 52, 6735–6740.
Couzin-Frankel, J. (2010). Bacteria and asthma: untangling the links. Science 330, 1168–1169. doi: 10.1126/science.330.6008.1168
Dixon, M. F., Genta, R. M., Yardley, J. H., and Correa, P. (1996). Classification and grading of gastritis. The updated Sydney System. International Workshop on the Histopathology of Gastritis, Houston 1994. Am. J. Surg. Pathol. 20, 1161–1181. doi: 10.1097/00000478-199610000-00001
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Eun, C. S., Kim, B. K., Han, D. S., Kim, S. Y., Kim, K. M., Choi, B. Y., et al. (2014). Differences in gastric mucosal microbiota profiling in patients with chronic gastritis, intestinal metaplasia, and gastric cancer using pyrosequencing methods. Helicobacter 19, 407–416. doi: 10.1111/hel.12145
Ferreira, R. M., Pereira-Marques, J., Pinto-Ribeiro, I., Costa, J. L., Carneiro, F., Machado, J. C., et al. (2018). Gastric microbial community profiling reveals a dysbiotic cancer-associated microbiota. Gut 67, 226–236. doi: 10.1136/gutjnl-2017-314205
Flemer, B., Lynch, D. B., Brown, J. M., Jeffery, I. B., Ryan, F. J., Claesson, M. J., et al. (2017). Tumour-associated and non-tumour-associated microbiota in colorectal cancer. Gut 66, 633–643. doi: 10.1136/gutjnl-2015-309595
Francino, M. P. (2015). Antibiotics and the human gut microbiome: dysbioses and accumulation of resistances. Front. Microbiol. 6:1543. doi: 10.3389/fmicb.2015.01543
Gopalakrishnan, V., Spencer, C. N., Nezi, L., Reuben, A., Andrews, M. C., Karpinets, T. V., et al. (2018). Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 359, 97–103. doi: 10.1126/science.aan4236
Heimesaat, M. M., Fischer, A., Plickert, R., Wiedemann, T., Loddenkemper, C., Gobel, U. B., et al. (2014). Helicobacter pylori induced gastric immunopathology is associated with distinct microbiota changes in the large intestines of long-term infected Mongolian gerbils. PLoS ONE 9:e100362. doi: 10.1371/journal.pone.0100362
Huws, S. A., Edwards, J. E., Kim, E. J., and Scollan, N. D. (2007). Specificity and sensitivity of eubacterial primers utilized for molecular profiling of bacteria within complex microbial ecosystems. J. Microbiol. Methods 70, 565–569. doi: 10.1016/j.mimet.2007.06.013
Jo, H. J., Kim, J., Kim, N., Park, J. H., Nam, R. H., Seok, Y. J., et al. (2016). Analysis of gastric microbiota by pyrosequencing: minor role of bacteria other than Helicobacter pylori in the gastric carcinogenesis. Helicobacter 21, 364–374. doi: 10.1111/hel.12293
Lertpiriyapong, K., Whary, M. T., Muthupalani, S., Lofgren, J. L., Gamazon, E. R., Feng, Y., et al. (2014). Gastric colonisation with a restricted commensal microbiota replicates the promotion of neoplastic lesions by diverse intestinal microbiota in the Helicobacter pylori INS-GAS mouse model of gastric carcinogenesis. Gut 63, 54–63. doi: 10.1136/gutjnl-2013-305178
Ley, R. E., Turnbaugh, P. J., Klein, S., and Gordon, J. I. (2006). Microbial ecology: human gut microbes associated with obesity. Nature 444, 1022–1023. doi: 10.1038/4441022a
Li, T. H., Qin, Y. W., Sham, P. C., Lau, K. S., Chu, K. M., and Leung, W. K. (2017). Alterations in gastric microbiota after H. pylori eradication and in different histological stages of gastric carcinogenesis. Sci. Rep. 7:44935. doi: 10.1038/srep44935
Lofgren, J. L., Whary, M. T., Ge, Z., Muthupalani, S., Taylor, N. S., Mobley, M., et al. (2011). Lack of commensal flora in Helicobacter pylori-infected INS-GAS mice reduces gastritis and delays intraepithelial neoplasia. Gastroenterology 140, 210–220. doi: 10.1053/j.gastro.2010.09.048
Magoc, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Maldonado-Contreras, A., Goldfarb, K. C., Godoy-Vitorino, F., Karaoz, U., Contreras, M., Blaser, M. J., et al. (2011). Structure of the human gastric bacterial community in relation to Helicobacter pylori status. ISME J. 5, 574–579. doi: 10.1038/ismej.2010.149
Mortha, A., Chudnovskiy, A., Hashimoto, D., Bogunovic, M., Spencer, S. P., Belkaid, Y., et al. (2014). Microbiota-dependent crosstalk between macrophages and ILC3 promotes intestinal homeostasis. Science 343:1249288. doi: 10.1126/science.1249288
Myllyluoma, E., Ahlroos, T., Veijola, L., Rautelin, H., Tynkkynen, S., and Korpela, R. (2007). Effects of anti-Helicobacter pylori treatment and probiotic supplementation on intestinal microbiota. Int. J. Antimicrob. Agents 29, 66–72. doi: 10.1016/j.ijantimicag.2006.08.034
Pan, K. F., Zhang, L., Gerhard, M., Ma, J. L., Liu, W. D., Ulm, K., et al. (2016). A large randomised controlled intervention trial to prevent gastric cancer by eradication of Helicobacter pylori in Linqu County, China: baseline results and factors affecting the eradication. Gut 65, 9–18. doi: 10.1136/gutjnl-2015-309197
Parks, D. H., Tyson, G. W., Hugenholtz, P., and Beiko, R. G. (2014). STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 30, 3123–3124. doi: 10.1093/bioinformatics/btu494
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Rakoff-Nahoum, S., and Medzhitov, R. (2009). Toll-like receptors and cancer. Nat. Rev. Cancer 9, 57–63. doi: 10.1038/nrc2541
Rooks, M. G., and Garrett, W. S. (2016). Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 16, 341–352. doi: 10.1038/nri.2016.42
Schulz, C., Schütte, K., Koch, N., Vilchez-Vargas, R., Wos-Oxley, M. L., Oxley, A. P. A., et al. (2018). The active bacterial assemblages of the upper GI tract in individuals with and without Helicobacter infection. Gut 67, 216–225. doi: 10.1136/gutjnl-2016-312904
Sekirov, I., Russell, S. L., Antunes, L. C., and Finlay, B. B. (2010). Gut microbiota in health and disease. Physiol. Rev. 90, 859–904. doi: 10.1152/physrev.00045.2009
Tang, R., Wei, Y., Li, Y., Chen, W., Chen, H., Wang, Q., et al. (2018). Gut microbial profile is altered in primary biliary cholangitis and partially restored after UDCA therapy. Gut 67, 534–541. doi: 10.1136/gutjnl-2016-313332
Torre, L. A., Bray, F., Siegel, R. L., Ferlay, J., Lortet-Tieulent, J., and Jemal, A. (2015). Global cancer statistics, (2012). CA Cancer J. Clin. 65, 87–108. doi: 10.3322/caac.21262
Tözün, N., and Vardareli, E. (2016). Gut microbiome and gastrointestinal cancer: les liaisons dangereuses. J. Clin. Gastroenterol. 50(Suppl. 2), S191–S196. doi: 10.1097/MCG.0000000000000714
Wang, L., Zhou, J., Xin, Y., Geng, C., Tian, Z., Yu, X., et al. (2016). Bacterial overgrowth and diversification of microbiota in gastric cancer. Eur. J. Gastroenterol. Hepatol. 28, 261–266. doi: 10.1097/MEG.0000000000000542
Wong, B. C., Lam, S. K., Wong, W. M., Chen, J. S., Zheng, T. T., Feng, R. E., et al. (2004). Helicobacter pylori eradication to prevent gastric cancer in a high-risk region of China: a randomized controlled trial. JAMA 291, 187–194. doi: 10.1001/jama.291.2.187
Yap, T. W., Gan, H. M., Lee, Y. P., Leow, A. H., Azmi, A. N., Francois, F., et al. (2016). Helicobacter pylori eradication causes perturbation of the human gut microbiome in young adults. PLoS ONE 11:e0151893. doi: 10.1371/journal.pone.0151893
Keywords: Helicobacter pylori, gastric lesions, gut microbiota, microbial diversity, 16S ribosomal RNA gene sequencing
Citation: Gao J-J, Zhang Y, Gerhard M, Mejias-Luque R, Zhang L, Vieth M, Ma J-L, Bajbouj M, Suchanek S, Liu W-D, Ulm K, Quante M, Li Z-X, Zhou T, Schmid R, Classen M, Li W-Q, You W-C and Pan K-F (2018) Association Between Gut Microbiota and Helicobacter pylori-Related Gastric Lesions in a High-Risk Population of Gastric Cancer. Front. Cell. Infect. Microbiol. 8:202. doi: 10.3389/fcimb.2018.00202
Received: 02 March 2018; Accepted: 30 May 2018;
Published: 19 June 2018.
Edited by:
D. Scott Merrell, Uniformed Services University, United StatesReviewed by:
Jennifer McMillan Noto, Vanderbilt University Medical Center, United StatesVirginia Helena Albarracín, Center for Electron Microscopy (CIME), Argentina
Guoqin Yu, National Cancer Institute (NCI), United States
Copyright © 2018 Gao, Zhang, Gerhard, Mejias-Luque, Zhang, Vieth, Ma, Bajbouj, Suchanek, Liu, Ulm, Quante, Li, Zhou, Schmid, Classen, Li, You and Pan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei-Cheng You, weichengyou@yahoo.com
Kai-Feng Pan, pankaifeng2002@yahoo.com
†These authors have contributed equally to this work.
 Juan-Juan Gao
Juan-Juan Gao Yang Zhang1†
Yang Zhang1†  Markus Gerhard
Markus Gerhard Raquel Mejias-Luque
Raquel Mejias-Luque Michael Vieth
Michael Vieth