Abstract
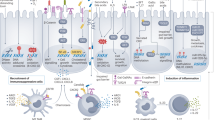
Colorectal cancer (CRC) accounts for about 10% of all new cancer cases globally. Located at close proximity to the colorectal epithelium, the gut microbiota comprises a large population of microorganisms that interact with host cells to regulate many physiological processes, such as energy harvest, metabolism and immune response. Sequencing studies have revealed microbial compositional and ecological changes in patients with CRC, whereas functional studies in animal models have pinpointed the roles of several bacteria in colorectal carcinogenesis, including Fusobacterium nucleatum and certain strains of Escherichia coli and Bacteroides fragilis. These findings give new opportunities to take advantage of our knowledge on the gut microbiota for clinical applications, such as gut microbiota analysis as screening, prognostic or predictive biomarkers, or modulating microorganisms to prevent cancer, augment therapies and reduce adverse effects of treatment. This Review aims to provide an overview and discussion of the gut microbiota in colorectal neoplasia, including relevant mechanisms in microbiota-related carcinogenesis, the potential of utilizing the microbiota as CRC biomarkers, and the prospect for modulating the microbiota for CRC prevention or treatment. These scientific findings will pave the way to clinically translate the use of gut microbiota for CRC in the near future.
Key points
-
Colorectal cancer (CRC) is one of the most common cancers; globally, it ranks third in incidence and second in mortality among all cancers.
-
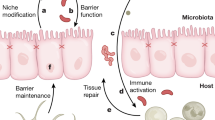
The gut microbiota comprises a large population of microorganisms that interact closely with host intestinal cells, and it can affect the immunity and metabolome in the gastrointestinal tract.
-
According to experimental evidence, the gut microbiota is involved in CRC formation, progression and its response to treatment.
-
Substantial changes in abundance of specific bacteria can be detected in patients with CRC and might serve as biomarkers for disease screening, prognostication and prediction of treatment response.
-
Modulation of the gut microbiota is a promising strategy to enhance treatment efficacy and reduce adverse effects of CRC therapies.
-
Future research should look into the best ways to modulate the gut microbiota and to investigate its short-term and long-term benefits through clinical trials.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Bray, F. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424 (2018).
Lichtenstein, P. et al. Environmental and heritable factors in the causation of cancer — analyses of cohorts of twins from Sweden, Denmark, and Finland. N. Engl. J. Med. 343, 78–85 (2000).
Foulkes, W. D. Inherited susceptibility to common cancers. N. Engl. J. Med. 359, 2143–2153 (2008).
Czene, K., Lichtenstein, P. & Hemminki, K. Environmental and heritable causes of cancer among 9.6 million individuals in the Swedish family-cancer database. Int. J. Cancer 99, 260–266 (2002).
Plummer, M. et al. Global burden of cancers attributable to infections in 2012: a synthetic analysis. Lancet Glob. Health 4, e609-e616 (2016).
Helmink, B. A., Khan, M. A. W., Hermann, A., Gopalakrishnan, V. & Wargo, J. A. The microbiome, cancer, and cancer therapy. Nat. Med. 25, 377–388 (2019).
Qin, J. et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 (2010).
Turnbaugh, P. J. et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031 (2006).
Chung, H. et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell 149, 1578–1593 (2012).
Brown, J. M. & Hazen, S. L. Microbial modulation of cardiovascular disease. Nat. Rev. Microbiol. 16, 171–181 (2018).
Maruvada, P., Leone, V., Kaplan, L. M. & Chang, E. B. The human microbiome and obesity: moving beyond associations. Cell Host Microbe 22, 589–599 (2017).
Sharon, G., Sampson, T. R., Geschwind, D. H. & Mazmanian, S. K. The central nervous system and the gut microbiome. Cell 167, 915–932 (2016).
Kostic, A. D., Xavier, R. J. & Gevers, D. The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology 146, 1489–1499 (2014).
Buffie, C. G. et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517, 205–208 (2015).
Laqueur, G. L., McDaniel, E. G. & Matsumoto, H. Tumor induction in germfree rats with methylazoxymethanol (MAM) and synthetic MAM acetate. J. Natl Cancer Inst. 39, 355–371 (1967).
Reddy, B. S., Weisburger, J. H., Narisawa, T. & Wynder, E. L. Colon carcinogenesis in germ-free rats with 1,2-dimethylhydrazine and N-methyl-n’-nitro-N-nitrosoguanidine. Cancer Res. 34, 2368–2372 (1974).
Reddy, B. S. et al. Colon carcinogenesis with azoxymethane and dimethylhydrazine in germ-free rats. Cancer Res. 35, 287–290 (1975). This study provides early evidence that gut microbiota can alter the effect of carcinogens in the large intestine.
Onoue, M., Kado, S., Sakaitani, Y., Uchida, K. & Morotomi, M. Specific species of intestinal bacteria influence the induction of aberrant crypt foci by 1,2-dimethylhydrazine in rats. Cancer Lett. 113, 179–186 (1997).
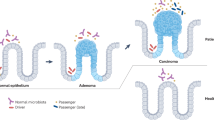
Wong, S. H. et al. Gavage of fecal samples from patients with colorectal cancer promotes intestinal carcinogenesis in germ-free and conventional mice. Gastroenterology 153, 1621–1633 e1626 (2017). Using faecal microbiota transplantation, this study demonstrates the carcinogenic properties of microbial communities obtained from patients with CRC in two mouse models.
Castellarin, M. et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 22, 299–306 (2012). This study reports the association between the abundance of Fusobacterium nucleatum and CRC in humans.
Feng, Q. et al. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat. Commun. 6, 6528 (2015).
Yu, J. et al. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut 66, 70–78 (2017).
Sobhani, I. et al. Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS One 6, (e16393 (2011).
Wang, T. et al. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 6, 320–329 (2012).
Ahn, J. et al. Human gut microbiome and risk for colorectal cancer. J. Natl Cancer Inst. 105, 1907–1911 (2013).
Zeller, G. et al. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol. Syst. Biol. 10, 766 (2014).
Zackular, J. P., Rogers, M. A., Ruffin, M. Tt & Schloss, P. D. The human gut microbiome as a screening tool for colorectal cancer. Cancer Prev. Res. (Phila) 7, 1112–1121 (2014).
Baxter, N. T., Ruffin, M. Tt, Rogers, M. A. & Schloss, P. D. Microbiota-based model improves the sensitivity of fecal immunochemical test for detecting colonic lesions. Genome Med. 8, 37 (2016).
Yazici, C. et al. Race-dependent association of sulfidogenic bacteria with colorectal cancer. Gut 66, 1983–1994 (2017).
Thomas, A. M. et al. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat. Med. 25, 667–678 (2019).
Wirbel, J. et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat. Med. 25, 679–689 (2019).
Yachida, S. et al. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat. Med. 25, 968–976 (2019).
Marchesi, J. R. et al. Towards the human colorectal cancer microbiome. PLoS One 6, e20447 (2011).
Chen, W., Liu, F., Ling, Z., Tong, X. & Xiang, C. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS One 7, e39743 (2012).
Kostic, A. D. et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 14, 207–215 (2013). This study evaluates the role of F. nucleatum in CRC, providing support that it could generate a pro-inflammatory microenvironment conducive to the progression of colorectal neoplasma.
Warren, R. L. et al. Co-occurrence of anaerobic bacteria in colorectal carcinomas. Microbiome 1, 16 (2013).
Zhang, Z. et al. Spatial heterogeneity and co-occurrence patterns of human mucosal-associated intestinal microbiota. ISME J. 8, 881–893 (2014).
Allali, I. et al. Gut microbiome compositional and functional differences between tumor and non-tumor adjacent tissues from cohorts from the US and Spain. Gut Microbes 6, 161–172 (2015).
Nakatsu, G. et al. Gut mucosal microbiome across stages of colorectal carcinogenesis. Nat. Commun. 6, 8727 (2015). This study depicts the mucosal microbiota landscape along different stages of colorectal carcinogenesis and reports changes in bacterial abundance and interspecies interactions in patients with colorectal adenoma and CRC.
Flemer, B. et al. Tumour-associated and non-tumour-associated microbiota in colorectal cancer. Gut 66, 633–643 (2017).
Wu, S. et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat. Med. 15, 1016–1022 (2009). This study establishes enterotoxigenic Bacteroides fragilis as a cancer-promoting bacterium via activation of the T H 17 cell response.
Arthur, J. C. et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 338, 120–123 (2012). Using a mouse model of colitis, this landmark study establishes genotoxic E. coli as a cancer-inducing bacterium in CRC.
Huycke, M. M., Abrams, V. & Moore, D. R. Enterococcus faecalis produces extracellular superoxide and hydrogen peroxide that damages colonic epithelial cell DNA. Carcinogenesis 23, 529–536 (2002).
Boleij, A. & Tjalsma, H. The itinerary of Streptococcus gallolyticus infection in patients with colonic malignant disease. Lancet Infect. Dis. 13, 719–724 (2013).
Zhernakova, A. et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science 352, 565–569 (2016).
He, Y. et al. Regional variation limits applications of healthy gut microbiome reference ranges and disease models. Nat. Med. 24, 1532–1535 (2018).
Drewes, J. L. et al. High-resolution bacterial 16S rRNA gene profile meta-analysis and biofilm status reveal common colorectal cancer consortia. NPJ Biofilms Microbiomes 3, 34 (2017).
Dai, Z. et al. Multi-cohort analysis of colorectal cancer metagenome identified altered bacteria across populations and universal bacterial markers. Microbiome 6, 70 (2018).
Shah, M. S. et al. Leveraging sequence-based faecal microbial community survey data to identify a composite biomarker for colorectal cancer. Gut 67, 882–891 (2018).
Dai, Z., Wong, S. H., Yu, J. & Wei, Y. Batch effects correction for microbiome data with Dirichlet-multinomial regression. Bioinformatics 35, 2348 (2018).
Sears, C. L., Geis, A. L. & Housseau, F. Bacteroides fragilis subverts mucosal biology: from symbiont to colon carcinogenesis. J. Clin. Invest. 124, 4166–4172 (2014).
Chung, L. et al. Bacteroides fragilis toxin coordinates a pro-carcinogenic inflammatory cascade via targeting of colonic epithelial cells. Cell Host Microbe 23, 421 (2018).
Seki, H. et al. Prevention of antibiotic-associated diarrhea in children by Clostridium butyricum MIYAIRI. Pediatr. Int. 45, 86–90 (2003).
Correa, N. B., Peret Filho, L. A., Penna, F. J., Lima, F. M. & Nicoli, J. R. A randomized formula controlled trial of Bifidobacterium lactis and Streptococcus thermophilus for prevention of antibiotic-associated diarrhea in infants. J. Clin. Gastroenterol. 39, 385–389 (2005).
Collins, D., Hogan, A. M. & Winter, D. C. Microbial and viral pathogens in colorectal cancer. Lancet Oncol. 12, 504–512 (2011).
Harkins, L. et al. Specific localisation of human cytomegalovirus nucleic acids and proteins in human colorectal cancer. Lancet 360, 1557–1563 (2002).
Bender, C. et al. Analysis of colorectal cancers for human cytomegalovirus presence. Infect. Agent Cancer 4, 6 (2009).
Laghi, L. et al. JC virus DNA is present in the mucosa of the human colon and in colorectal cancers. Proc Natl Acad Sci USA 96, 7484–7489 (1999).
Cheng, J. Y., Sheu, L. F., Meng, C. L., Lee, W. H. & Lin, J. C. Detection of human papillomavirus DNA in colorectal carcinomas by polymerase chain reaction. Gut 37, 87–90 (1995).
Hart, H., Neill, W. A. & Norval, M. Lack of association of cytomegalovirus with adenocarcinoma of the colon. Gut 23, 21–30 (1982).
Knosel, T., Schewe, C., Dietel, M. & Petersen, I. Cytomegalovirus is not associated with progression and metastasis of colorectal cancer. Cancer Lett. 211, 243–247 (2004).
Gornick, M. C. et al. Human papillomavirus is not associated with colorectal cancer in a large international study. Cancer Causes Control 21, 737–743 (2010).
Nakatsu, G. et al. Alterations in enteric virome are associated with colorectal cancer and survival outcomes. Gastroenterology 155, 529–541.e5 (2018). This study is the first to report the gut virome in patients with CRC and identifies viral markers that could classify cases and controls.
Hannigan, G. D., Duhaime, M. B., Ruffin, M. Tt, Koumpouras, C. C. & Schloss, P. D. Diagnostic potential and interactive dynamics of the colorectal cancer virome. mBio 9, e02248-18 (2018).
Gao, R. et al. Dysbiosis signature of mycobiota in colon polyp and colorectal cancer. Eur. J. Clin. Microbiol Infect. Dis. 36, 2457–2468 (2017).
Coker, O. O. et al. Enteric fungal microbiota dysbiosis and ecological alterations in colorectal cancer. Gut 68, 654–662 (2019).
Strum, W. B. Colorectal adenomas. N. Engl. J. Med. 374, 1065–1075 (2016).
Shen, X. J. et al. Molecular characterization of mucosal adherent bacteria and associations with colorectal adenomas. Gut Microbes 1, 138–147 (2010).
Sanapareddy, N. et al. Increased rectal microbial richness is associated with the presence of colorectal adenomas in humans. ISME J. 6, 1858–1868 (2012).
McCoy, A. N. et al. Fusobacterium is associated with colorectal adenomas. PLOS ONE 8, e53653 (2013).
Lu, Y. et al. Mucosal adherent bacterial dysbiosis in patients with colorectal adenomas. Sci. Rep. 6, 26337 (2016).
Luan, C. et al. Dysbiosis of fungal microbiota in the intestinal mucosa of patients with colorectal adenomas. Sci. Rep. 5, 7980 (2015).
Tilg, H., Adolph, T. E., Gerner, R. R. & Moschen, A. R. The intestinal microbiota in colorectal cancer. Cancer Cell 33, 954–964 (2018).
Round, J. L. & Mazmanian, S. K. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 9, 313–323 (2009).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Lasry, A., Zinger, A. & Ben-Neriah, Y. Inflammatory networks underlying colorectal cancer. Nat. Immunol. 17, 230–240 (2016).
Beaugerie, L. & Itzkowitz, S. H. Cancers complicating inflammatory bowel disease. N. Engl. J. Med. 373, 195 (2015).
Eaden, J. A., Abrams, K. R. & Mayberry, J. F. The risk of colorectal cancer in ulcerative colitis: a meta-analysis. Gut 48, 526–535 (2001).
Canavan, C., Abrams, K. R. & Mayberry, J. Meta-analysis: colorectal and small bowel cancer risk in patients with Crohn’s disease. Aliment. Pharmacol. Ther. 23, 1097–1104 (2006).
Sebastian, S. et al. Colorectal cancer in inflammatory bowel disease: results of the 3rd ECCO pathogenesis scientific workshop (I). J. Crohns Colitis 8, 5–18 (2014).
Cremonesi, E. et al. Gut microbiota modulate T cell trafficking into human colorectal cancer. Gut 67, 1984–1994 (2018).
Tomkovich, S. et al. Locoregional effects of microbiota in a preclinical model of colon carcinogenesis. Cancer Res. 77, 2620–2632 (2017).
Boleij, A. et al. The Bacteroides fragilistoxin gene is prevalent in the colon mucosa of colorectal cancer patients. Clin. Infect. Dis. 60, 208–215 (2015).
Arthur, J. C. et al. Microbial genomic analysis reveals the essential role of inflammation in bacteria-induced colorectal cancer. Nat. Commun. 5, 4724 (2014).
Bonnet, M. et al. Colonization of the human gut by E. coli and colorectal cancer risk. Clin. Cancer Res. 20, 859–867 (2014).
Wang, X. et al. 4-hydroxy-2-nonenal mediates genotoxicity and bystander effects caused by Enterococcus faecalis-infected macrophages. Gastroenterology 142, 543–551 e547 (2012).
Moschen, A. R. et al. Lipocalin 2 protects from inflammation and tumorigenesis associated with gut microbiota alterations. Cell Host Microbe 19, 455–469 (2016).
Kesselring, R. et al. IRAK-M expression in tumor cells supports colorectal cancer progression through reduction of antimicrobial defense and stabilization of STAT3. Cancer Cell 29, 684–696 (2016).
Couturier-Maillard, A. et al. NOD2-mediated dysbiosis predisposes mice to transmissible colitis and colorectal cancer. J. Clin. Invest. 123, 700–711 (2013).
Zhu, H. et al. RNA virus receptor Rig-I monitors gut microbiota and inhibits colitis-associated colorectal cancer. J. Exp. Clin. Cancer Res. 36, 2 (2017).
Man, S. M. et al. Critical role for the DNA sensor AIM2 in stem cell proliferation and cancer. Cell 162, 45–58 (2015).
Yang, Y. et al. Fusobacterium nucleatum increases proliferation of colorectal cancer cells and tumor development in mice by activating toll-like receptor 4 signaling to nuclear factor-kappab, and up-regulating expression of microRNA-21. Gastroenterology 152, 851–866 e824 (2017).
Wu, Y. et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis in mice via a toll-like receptor 4/p21-activated kinase 1 cascade. Dig. Dis. Sci. 63, 1210–1218 (2018).
Tsoi, H. et al. Peptostreptococcus anaerobius induces intracellular cholesterol biosynthesis in colon cells to induce proliferation and causes dysplasia in mice. Gastroenterology 152, 1419–1433 e1415 (2017).
Doll, R. & Peto, R. The causes of cancer: quantitative estimates of avoidable risks of cancer in the United States today. J. Natl Cancer Inst. 66, 1191–1308 (1981).
Zhang, F. F. et al. Preventable cancer burden associated with poor diet in the United States. JNCI Cancer Spectr. 3, pkz034 (2019).
Alexander, D. D., Weed, D. L., Cushing, C. A. & Lowe, K. A. Meta-analysis of prospective studies of red meat consumption and colorectal cancer. Eur. J. Cancer Prev. 20, 293–307 (2011).
O’Keefe, S. J. Diet, microorganisms and their metabolites, and colon cancer. Nat. Rev. Gastroenterol. Hepatol. 13, 691–706 (2016).
Louis, P., Hold, G. L. & Flint, H. J. The gut microbiota, bacterial metabolites and colorectal cancer. Nat. Rev. Microbiol. 12, 661–672 (2014).
Ijssennagger, N. et al. Gut microbiota facilitates dietary heme-induced epithelial hyperproliferation by opening the mucus barrier in colon. Proc. Natl Acad. Sci. USA 112, 10038–10043 (2015).
Singh, N. et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 40, 128–139 (2014).
Chang, P. V., Hao, L., Offermanns, S. & Medzhitov, R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc. Natl Acad. Sci. USA 111, 2247–2252 (2014).
Buda, A. et al. Butyrate downregulates alpha2beta1 integrin: a possible role in the induction of apoptosis in colorectal cancer cell lines. Gut 52, 729–734 (2003).
Furusawa, Y. et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504, 446–450 (2013).
Smith, P. M. et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341, 569–573 (2013).
Ou, J. et al. Diet, microbiota, and microbial metabolites in colon cancer risk in rural Africans and African Americans. Am. J. Clin. Nutr. 98, 111–120 (2013).
Vernia, P., Gnaedinger, A., Hauck, W. & Breuer, R. I. Organic anions and the diarrhea of inflammatory bowel disease. Dig. Dis. Sci. 33, 1353–1358 (1988).
Chen, H. M. et al. Decreased dietary fiber intake and structural alteration of gut microbiota in patients with advanced colorectal adenoma. Am. J. Clin. Nutr. 97, 1044–1052 (2013).
Bultman, S. J. Molecular pathways: gene-environment interactions regulating dietary fiber induction of proliferation and apoptosis via butyrate for cancer prevention. Clin. Cancer Res. 20, 799–803 (2014).
Belcheva, A. et al. Gut microbial metabolism drives transformation of MSH2-deficient colon epithelial cells. Cell 158, 288–299 (2014).
Bultman, S. J. & Jobin, C. Microbial-derived butyrate: an oncometabolite or tumor-suppressive metabolite? Cell Host Microbe 16, 143–145 (2014).
de Aguiar Vallim, T. Q., Tarling, E. J. & Edwards, P. A. Pleiotropic roles of bile acids in metabolism. Cell Metab. 17, 657–669 (2013).
Ou, J., DeLany, J. P., Zhang, M., Sharma, S. & O’Keefe, S. J. Association between low colonic short-chain fatty acids and high bile acids in high colon cancer risk populations. Nutr. Cancer 64, 34–40 (2012).
Chomchai, C., Bhadrachari, N. & Nigro, N. D. The effect of bile on the induction of experimental intestinal tumors in rats. Dis. Colon Rectum 17, 310–312 (1974).
Bernstein, H., Bernstein, C., Payne, C. M. & Dvorak, K. Bile acids as endogenous etiologic agents in gastrointestinal cancer. World J. Gastroenterol. 15, 3329–3340 (2009).
Bernstein, C. et al. Carcinogenicity of deoxycholate, a secondary bile acid. Arch. Toxicol. 85, 863–871 (2011).
Cuevas-Ramos, G. et al. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc. Natl Acad. Sci. USA 107, 11537–11542 (2010).
He, Z. et al. Campylobacter jejuni promotes colorectal tumorigenesis through the action of cytolethal distending toxin. Gut 68, 289–300 (2019).
Shen, Z. et al. Cytolethal distending toxin promotes Helicobacter cinaedi-associated typhlocolitis in interleukin-10-deficient mice. Infect. Immun. 77, 2508–2516 (2009).
Buc, E. et al. High prevalence of mucosa-associated E. coli producing cyclomodulin and genotoxin in colon cancer. PLOS ONE 8, e56964 (2013).
Goodwin, A. C. et al. Polyamine catabolism contributes to enterotoxigenic Bacteroides fragilis-induced colon tumorigenesis. Proc. Natl Acad. Sci. USA 108, 15354–15359 (2011).
Wang, X. & Huycke, M. M. Extracellular superoxide production by Enterococcus faecalis promotes chromosomal instability in mammalian cells. Gastroenterology 132, 551–561 (2007).
Cougnoux, A. et al. Small-molecule inhibitors prevent the genotoxic and protumoural effects induced by colibactin-producing bacteria. Gut 65, 278–285 (2016).
Tjalsma, H., Boleij, A., Marchesi, J. R. & Dutilh, B. E. A bacterial driver-passenger model for colorectal cancer: beyond the usual suspects. Nat. Rev. Microbiol. 10, 575–582 (2012). This Perspective proposes the driver–passenger carcinogenesis model in which driver and passenger bacteria have distinct roles in the tissue microenvironment.
Lauby-Secretan, B. et al. The IARC perspective on colorectal cancer screening. New Engl. J. Med. 378, 1734–1740 (2018).
Siegel, R. L., Miller, K. D. & Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 69, 7–34 (2019).
Lee, J. K., Liles, E. G., Bent, S., Levin, T. R. & Corley, D. A. Accuracy of fecal immunochemical tests for colorectal cancer: systematic review and meta-analysis. Ann. Intern. Med. 160, 171 (2014).
Hundt, S., Haug, U. & Brenner, H. Comparative evaluation of immunochemical fecal occult blood tests for colorectal adenoma detection. Ann. Intern. Med. 150, 162–169 (2009).
Imperiale, T. F. et al. Multitarget stool DNA testing for colorectal-cancer screening. N. Engl. J. Med. 370, 1287–1297 (2014).
Wong, S. H. et al. Quantitation of faecal Fusobacterium improves faecal immunochemical test in detecting advanced colorectal neoplasia. Gut 66, 1441–1448 (2017). This study reports that quantifying faecal Fusobacterium could improve the performance of the occult blood test in detecting CRC and advanced adenoma.
Suehiro, Y. et al. Highly sensitive faecal DNA testing of TWIST1 methylation in combination with faecal immunochemical test for haemoglobin is a promising marker for detection of colorectal neoplasia. Ann. Clin. Biochem. 55, 59–68 (2018).
Guo, S. et al. A simple and novel fecal biomarker for colorectal cancer: ratio of Fusobacterium nucleatum to probiotics populations, based on their antagonistic effect. Clin. Chem. 64, 1327–1337 (2018).
Xie, Y. H. et al. Fecal clostridium symbiosum for noninvasive detection of early and advanced colorectal cancer: test and validation studies. EBioMedicine 25, 32–40 (2017).
Eklof, V. et al. Cancer-associated fecal microbial markers in colorectal cancer detection. Int. J. Cancer 141, 2528–2536 (2017).
Liang, Q. et al. Fecal bacteria act as novel biomarkers for noninvasive diagnosis of colorectal cancer. Clin Cancer Res. 23, 2061–2070 (2017).
Fearon, E. R. & Vogelstein, B. A genetic model for colorectal tumorigenesis. Cell 61, 759–767 (1990).
Rezasoltani, S. et al. The association between fecal microbiota and different types of colorectal polyp as precursors of colorectal cancer. Microb. Pathog. 124, 244–249 (2018).
Flanagan, L. et al. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur. J. Clin. Microbiol Infect. Dis. 33, 1381–1390 (2014).
Yu, J. et al. Invasive Fusobacterium nucleatum may play a role in the carcinogenesis of proximal colon cancer through the serrated neoplasia pathway. Int. J. Cancer 139, 1318–1326 (2016).
Russo, E. et al. Preliminary comparison of oral and intestinal human microbiota in patients with colorectal cancer: a pilot study. Front. Microbiol. 8, 2699 (2017).
Flemer, B. et al. The oral microbiota in colorectal cancer is distinctive and predictive. Gut 67, 1454–1463 (2018).
Corredoira-Sanchez, J. et al. Association between bacteremia due to Streptococcus gallolyticus subsp. gallolyticus (Streptococcus bovis I) and colorectal neoplasia: a case-control study. Clin. Infect. Dis. 55, 491–496 (2012).
Kwong, T. N. Y. et al. Association between bacteremia from specific microbes and subsequent diagnosis of colorectal cancer. Gastroenterology 155, 383–390.e8 (2018).
Butt, J. et al. Prospective evaluation of antibody response to Streptococcus gallolyticus and risk of colorectal cancer. Int. J. Cancer 143, 245–252 (2018).
Butt, J. et al. Antibody responses to Streptococcus gallolyticus subspecies gallolyticusproteins in a large prospective colorectal cancer cohort consortium. Cancer Epidemiol. Biomarkers Prev. 27, 1186–1194 (2018).
Wang, H. F. et al. Evaluation of antibody level against Fusobacterium nucleatum in the serological diagnosis of colorectal cancer. Sci. Rep. 6, 33440 (2016).
Weaver, G. A., Krause, J. A., Miller, T. L. & Wolin, M. J. Short chain fatty acid distributions of enema samples from a sigmoidoscopy population: an association of high acetate and low butyrate ratios with adenomatous polyps and colon cancer. Gut 29, 1539–1543 (1988).
Weir, T. L. et al. Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PLOS ONE 8, e70803 (2013).
Wang, X., Wang, J., Rao, B. & Deng, L. Gut flora profiling and fecal metabolite composition of colorectal cancer patients and healthy individuals. Exp. Ther. Med. 13, 2848–2854 (2017).
Mima, K. et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut 65, 1973–1980 (2016).
Wei, Z. et al. Could gut microbiota serve as prognostic biomarker associated with colorectal cancer patients’ survival? A pilot study on relevant mechanism. Oncotarget 7, 46158–46172 (2016).
Tahara, T. et al. Fusobacterium in colonic flora and molecular features of colorectal carcinoma. Cancer Res. 74, 1311–1318 (2014).
Ito, M. et al. Association of Fusobacterium nucleatum with clinical and molecular features in colorectal serrated pathway. Int. J. Cancer 137, 1258–1268 (2015).
Mima, K. et al. Fusobacterium nucleatum in colorectal carcinoma tissue according to tumor location. Clin. Transl Gastroenterol. 7, e200 (2016).
Kerr, J., Anderson, C. & Lippman, S. M. Physical activity, sedentary behaviour, diet, and cancer: an update and emerging new evidence. Lancet Oncol. 18, e457–e471 (2017).
Song, M. & Giovannucci, E. Preventable incidence and mortality of carcinoma associated with lifestyle factors among white adults in the United States. JAMA Oncol. 2, 1154–1161 (2016).
Islami, F. et al. Proportion and number of cancer cases and deaths attributable to potentially modifiable risk factors in the United States. CA Cancer J. Clin. 68, 31–54 (2018).
Carmody, R. N. et al. Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe 17, 72–84 (2015).
Rothschild, D. et al. Environment dominates over host genetics in shaping human gut microbiota. Nature 555, 210–215 (2018).
David, L. A. et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563 (2014).
O’Keefe, S. J. et al. Fat, fibre and cancer risk in African Americans and rural Africans. Nat. Commun. 6, 6342 (2015). This study shows that dietary change alone is sufficient to shift the microbiota, affect intestinal inflammation and alter cell proliferation markers that could reflect carcinogenesis.
De Filippo, C. et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl Acad. Sci. USA 107, 14691–14696 (2010).
Makki, K., Deehan, E. C., Walter, J. & Backhed, F. The impact of dietary fiber on gut microbiota in host health and disease. Cell Host Microbe 23, 705–715 (2018).
Gibson, G. R. et al. Expert consensus document: the International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat. Rev. Gastroenterol. Hepatol. 14, 491–502 (2017).
So, D. et al. Dietary fiber intervention on gut microbiota composition in healthy adults: a systematic review and meta-analysis. Am. J. Clin. Nutr. 107, 965–983 (2018).
Donohoe, D. R. et al. A gnotobiotic mouse model demonstrates that dietary fiber protects against colorectal tumorigenesis in a microbiota- and butyrate-dependent manner. Cancer Discov. 4, 1387–1397 (2014).
Alberts, D. S. et al. Lack of effect of a high-fiber cereal supplement on the recurrence of colorectal adenomas. Phoenix Colon Cancer Prevention Physicians’ Network. N. Engl. J. Med. 342, 1156–1162 (2000).
Schatzkin, A. et al. Lack of effect of a low-fat, high-fiber diet on the recurrence of colorectal adenomas. Polyp Prevention Trial Study Group. N. Engl. J. Med. 342, 1149–1155 (2000).
Lanza, E. et al. The Polyp Prevention trial continued follow-up study: no effect of a low-fat, high-fiber, high-fruit, and -vegetable diet on adenoma recurrence eight years after randomization. Cancer Epidemiol. Biomarkers Prev. 16, 1745–1752 (2007).
Chan, D. S. et al. Red and processed meat and colorectal cancer incidence: meta-analysis of prospective studies. PLOS ONE 6, e20456 (2011).
Bouvard, V. et al. Carcinogenicity of consumption of red and processed meat. Lancet Oncol. 16, 1599–1600 (2015).
Orlich, M. J. et al. Vegetarian dietary patterns and the risk of colorectal cancers. JAMA Intern. Med. 175, 767–776 (2015).
Hildebrandt, M. A. et al. High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology 137, 1716–1724.e1-2 (2009).
Devkota, S. et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10-/- mice. Nature 487, 104–108 (2012).
Zhang, C. et al. Structural resilience of the gut microbiota in adult mice under high-fat dietary perturbations. ISME J. 6, 1848–1857 (2012).
Cao, H. et al. The secondary bile acid, deoxycholate accelerates intestinal adenoma-adenocarcinoma sequence in Apcmin/+ mice through enhancing Wnt signaling. Fam. Cancer 13, 563–571 (2014).
Drasar, B. S. & Irving, D. Environmental factors and cancer of the colon and breast. Br. J. Cancer 27, 167–172 (1973).
Oba, S. et al. The relationship between the consumption of meat, fat, and coffee and the risk of colon cancer: a prospective study in Japan. Cancer Lett. 244, 260–267 (2006).
Liu, L. et al. Is dietary fat associated with the risk of colorectal cancer? A meta-analysis of 13 prospective cohort studies. Eur. J. Nutr. 50, 173–184 (2011).
MacLennan, R. et al. Randomized trial of intake of fat, fiber, and beta carotene to prevent colorectal adenomas. J. Natl Cancer Inst. 87, 1760–1766 (1995).
Thomson, C. A. et al. Cancer incidence and mortality during the intervention and postintervention periods of the Women’s Health Initiative dietary modification trial. Cancer Epidemiol. Biomarkers Prev. 23, 2924–2935 (2014).
Le Chatelier, E. et al. Richness of human gut microbiome correlates with metabolic markers. Nature 500, 541–546 (2013).
Ley, R. E. et al. Obesity alters gut microbial ecology. Proc. Natl Acad. Sci. USA 102, 11070–11075 (2005).
Ley, R. E., Turnbaugh, P. J., Klein, S. & Gordon, J. I. Microbial ecology: human gut microbes associated with obesity. Nature 444, 1022–1023 (2006).
Li, R. et al. Obesity, rather than diet, drives epigenomic alterations in colonic epithelium resembling cancer progression. Cell Metab. 19, 702–711 (2014).
Qin, Y. et al. An obesity-associated gut microbiome reprograms the intestinal epigenome and leads to altered colonic gene expression. Genome Biol. 19, 7 (2018).
Liu, R. et al. Gut microbiome and serum metabolome alterations in obesity and after weight-loss intervention. Nat. Med. 23, 859–868 (2017).
Seganfredo, F. B. et al. Weight-loss interventions and gut microbiota changes in overweight and obese patients: a systematic review. Obes. Rev. 18, 832–851 (2017).
Keum, N. et al. Adult weight gain and adiposity-related cancers: a dose-response meta-analysis of prospective observational studies. J. Natl Cancer Inst. 107, djv088 (2015).
Sjostrom, L. et al. Effects of bariatric surgery on cancer incidence in obese patients in Sweden (Swedish Obese Subjects Study): a prospective, controlled intervention trial. Lancet Oncol. 10, 653–662 (2009).
Derogar, M. et al. Increased risk of colorectal cancer after obesity surgery. Ann. Surg. 258, 983–988 (2013).
Aravani, A. et al. Obesity surgery and risk of colorectal and other obesity-related cancers: an English population-based cohort study. Cancer Epidemiol. 53, 99–104 (2018).
Schauer, D. P. et al. Bariatric surgery and the risk of cancer in a large multisite cohort. Ann. Surg. 269, 95–101 (2019).
Hull, M. A., Markar, S. R. & Morris, E. J. A. Cancer risk after bariatric surgery — is colorectal cancer a special case? Nat. Rev. Gastroenterol. Hepatol. 15, 653–654 (2018).
Hill, C. et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 11, 506–514 (2014).
Zitvogel, L., Daillere, R., Roberti, M. P., Routy, B. & Kroemer, G. Anticancer effects of the microbiome and its products. Nat. Rev. Microbiol. 15, 465–478 (2017).
Dos Reis, S. A. et al. Review of the mechanisms of probiotic actions in the prevention of colorectal cancer. Nutr. Res. 37, 1–19 (2017).
Thirabunyanon, M., Boonprasom, P. & Niamsup, P. Probiotic potential of lactic acid bacteria isolated from fermented dairy milks on antiproliferation of colon cancer cells. Biotechnol. Lett. 31, 571–576 (2009).
Chen, C. C. et al. Oral inoculation of probiotics Lactobacillus acidophilus NCFM suppresses tumour growth both in segmental orthotopic colon cancer and extra-intestinal tissue. Br. J. Nutr. 107, 1623–1634 (2012).
Wan, Y. et al. Fermentation supernatants of Lactobacillus delbrueckii inhibit growth of human colon cancer cells and induce apoptosis through a caspase 3-dependent pathway. Oncol. Lett. 7, 1738–1742 (2014).
Konishi, H. et al. Probiotic-derived ferrichrome inhibits colon cancer progression via JNK-mediated apoptosis. Nat. Commun. 7, 12365 (2016).
Corthesy, B., Gaskins, H. R. & Mercenier, A. Cross-talk between probiotic bacteria and the host immune system. J. Nutr. 137, 781S–790S (2007).
Delcenserie, V. et al. Immunomodulatory effects of probiotics in the intestinal tract. Curr. Issues Mol. Biol. 10, 37–54 (2008).
Burns, A. J. & Rowland, I. R. Antigenotoxicity of probiotics and prebiotics on faecal water-induced DNA damage in human colon adenocarcinoma cells. Mutat. Res. 551, 233–243 (2004).
Nowak, A. & Libudzisz, Z. Ability of probiotic Lactobacillus casei DN 114001 to bind or/and metabolise heterocyclic aromatic amines in vitro. Eur. J. Nutr. 48, 419–427 (2009).
Zhu, J. et al. Lactobacillus salivarius Ren prevent the early colorectal carcinogenesis in 1,2-dimethylhydrazine-induced rat model. J. Appl. Microbiol. 117, 208–216 (2014).
Ishikawa, H. et al. Randomized trial of dietary fiber and Lactobacillus caseiadministration for prevention of colorectal tumors. Int. J. Cancer 116, 762–767 (2005).
Rafter, J. et al. Dietary synbiotics reduce cancer risk factors in polypectomized and colon cancer patients. Am. J. Clin. Nutr. 85, 488–496 (2007).
Worthley, D. L. et al. A human, double-blind, placebo-controlled, crossover trial of prebiotic, probiotic, and synbiotic supplementation: effects on luminal, inflammatory, epigenetic, and epithelial biomarkers of colorectal cancer. Am. J. Clin. Nutr. 90, 578–586 (2009).
Zitvogel, L., Ma, Y., Raoult, D., Kroemer, G. & Gajewski, T. F. The microbiome in cancer immunotherapy: diagnostic tools and therapeutic strategies. Science 359, 1366–1370 (2018). This review discusses how gut microbiota can affect cancer immunotherapy and how this interaction could be utilized to enhance treatment efficacy and reduce adverse effects.
Garcia-Gonzalez, A. P. et al. Bacterial metabolism affects the C. elegansresponse to cancer chemotherapeutics. Cell 169, 431–441.e8 (2017).
Viaud, S. et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science 342, 971–976 (2013).
Daillere, R. et al. Enterococcus hiraeand Barnesiella intestinihominis facilitate cyclophosphamide-induced therapeutic immunomodulatory effects. Immunity 45, 931–943 (2016).
Vande Voorde, J. et al. Nucleoside-catabolizing enzymes in mycoplasma-infected tumor cell cultures compromise the cytostatic activity of the anticancer drug gemcitabine. J. Biol. Chem. 289, 13054–13065 (2014).
Iida, N. et al. Commensal bacteria control Cancer Res.ponse to therapy by modulating the tumor microenvironment. Science 342, 967–970 (2013).
Alexander, J. L. et al. Gut microbiota modulation of chemotherapy efficacy and toxicity. Nat. Rev. Gastroenterol. Hepatol. 14, 356–365 (2017).
Yu, T. et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell 170, 548–563 e516 (2017).
Wallace, B. D. et al. Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science 330, 831–835 (2010).
Wallace, B. D. et al. Structure and inhibition of microbiome beta-glucuronidases essential to the alleviation of cancer drug toxicity. Chem. Biol. 22, 1238–1249 (2015).
Yeung, C. Y. et al. Amelioration of chemotherapy-induced intestinal mucositis by orally administered probiotics in a mouse model. PLOS ONE 10, e0138746 (2015).
Kato, S. et al. Probiotic Bifidobacterium bifidum G9-1 attenuates 5-fluorouracil-induced intestinal mucositis in mice via suppression of dysbiosis-related secondary inflammatory responses. Clin. Exp. Pharmacol. Physiol. 44, 1017–1025 (2017).
Chang, C. W. et al. Lactobacillus casei variety rhamnosus probiotic preventively attenuates 5-fluorouracil/oxaliplatin-induced intestinal injury in a syngeneic colorectal cancer model. Front. Microbiol. 9, 983 (2018).
Wei, S. C., Duffy, C. R. & Allison, J. P. Fundamental mechanisms of immune checkpoint blockade therapy. Cancer Discov. 8, 1069–1086 (2018).
Routy, B. et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 359, 91–97 (2018).
Sivan, A. et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 350, 1084–1089 (2015).
Gopalakrishnan, V. et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 359, 97–103 (2018).
Matson, V. et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 359, 104–108 (2018).
Vetizou, M. et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 350, 1079–1084 (2015).
Peled, J. U. et al. Intestinal microbiota and relapse after hematopoietic-cell transplantation. J. Clin. Oncol. 35, 1650–1659 (2017).
Chaput, N. et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann. Oncol. 28, 1368–1379 (2017).
Gharaibeh, R. Z. & Jobin, C. Microbiota and cancer immunotherapy: in search of microbial signals. Gut https://doi.org/10.1136/gutjnl-2018-317220 (2018).
Tanoue, T. et al. A defined commensal consortium elicits CD8 T cells and anti-cancer immunity. Nature 565, 600–605 (2019).
Dubin, K. et al. Intestinal microbiome analyses identify melanoma patients at risk for checkpoint-blockade-induced colitis. Nat. Commun. 7, 10391 (2016).
Wang, Y. et al. Fecal microbiota transplantation for refractory immune checkpoint inhibitor-associated colitis. Nat. Med. 24, 1804–1808 (2018). This study reports for the first time that modifying gut microbiota by faecal transplantation can ameliorate refractory colitis as an adverse effect of immunotherapy.
Brahmer, J. R. et al. Phase I study of single-agent anti-programmed death-1 (MDX-1106) in refractory solid tumors: safety, clinical activity, pharmacodynamics, and immunologic correlates. J. Clin. Oncol. 28, 3167–3175 (2010).
Topalian, S. L. et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med. 366, 2443–2454 (2012).
Overman, M. J. et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): an open-label, multicentre, phase 2 study. Lancet Oncol. 18, 1182–1191 (2017).
Overman, M. J. et al. Durable clinical benefit with nivolumab plus ipilimumab in DNA mismatch repair-deficient/microsatellite instability-high metastatic colorectal cancer. J. Clin. Oncol. 36, 773–779 (2018).
Le, D. T. et al. PD-1 blockade in tumors with mismatch-repair deficiency. N. Engl. J. Med. 372, 2509–2520 (2015).
Le, D. T. et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357, 409–413 (2017).
Bullman, S. et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science 358, 1443–1448 (2017). This landmark study identifies the presence of Fusobacterium in metastatic CRC lesions and shows that oral metronidazole can shrink tumours in mice bearing a cancer xenograft.
D’Haens, G. R. & Jobin, C. Fecal microbial transplantation for diseases beyond recurrent Clostridium difficile infection. Gastroenterology 157, 624–636 (2019).
McQuade, J. L., Daniel, C. R., Helmink, B. A. & Wargo, J. A. Modulating the microbiome to improve therapeutic response in cancer. Lancet Oncol. 20, e77–e91 (2019).
Suez, J. & Elinav, E. The path towards microbiome-based metabolite treatment. Nat. Microbiol. 2, 17075 (2017).
Hu, Y. et al. Manipulation of the gut microbiota using resistant starch is associated with protection against colitis-associated colorectal cancer in rats. Carcinogenesis 37, 366–375 (2016).
Kaiko, G. E. et al. The colonic crypt protects stem cells from microbiota-derived metabolites. Cell 167, 1137 (2016).
Mimee, M., Citorik, R. J. & Lu, T. K. Microbiome therapeutics — advances and challenges. Adv. Drug Deliv. Rev. 105, 44–54 (2016).
Duan, F. & March, J. C. Engineered bacterial communication prevents Vibrio cholerae virulence in an infant mouse model. Proc. Natl Acad. Sci. USA 107, 11260–11264 (2010).
Steidler, L. et al. Treatment of murine colitis by Lactococcus lactis secreting interleukin-10. Science 289, 1352–1355 (2000).
Kommineni, S. et al. Bacteriocin production augments niche competition by enterococci in the mammalian gastrointestinal tract. Nature 526, 719–722 (2015).
Reardon, S. Phage therapy gets revitalized. Nature 510, 15–16 (2014).
Kingwell, K. Bacteriophage therapies re-enter clinical trials. Nat. Rev. Drug Discov. 14, 515–516 (2015).
Suez, J. et al. Post-antibiotic gut mucosal microbiome reconstitution is impaired by probiotics and improved by autologous FMT. Cell 174, 1406–1423 e1416 (2018).
Zmora, N. et al. Personalized gut mucosal colonization resistance to empiric probiotics is associated with unique host and microbiome features. Cell 174, 1388–1405 e1321 (2018).
van Nood, E. et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N. Engl. J. Med. 368, 407–415 (2013).
Smith, M. B., Kelly, C. & Alm, E. J. Policy: how to regulate faecal transplants. Nature 506, 290–291 (2014).
Sullivan, A., Edlund, C. & Nord, C. E. Effect of antimicrobial agents on the ecological balance of human microflora. Lancet Infect. Dis. 1, 101–114 (2001).
Dethlefsen, L. & Relman, D. A. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc. Natl Acad. Sci. USA 108, 4554–4561 (2011).
Koliaraki, V., Pallangyo, C. K., Greten, F. R. & Kollias, G. Mesenchymal cells in colon cancer. Gastroenterology 152, 964–979 (2017).
Manson McGuire, A. et al. Evolution of invasion in a diverse set of Fusobacteriumspecies. mBio 5, e01864 (2014).
Strauss, J. et al. Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm. Bowel Dis. 17, 1971–1978 (2011).
Brennan, C. A. & Garrett, W. S. Fusobacterium nucleatum – symbiont, opportunist and oncobacterium. Nat. Rev. Microbiol. 17, 156–166 (2019).
Flynn, K. J., Baxter, N. T. & Schloss, P. D. Metabolic and community synergy of oral bacteria in colorectal cancer. mSphere 1, e00102–e00116 (2016).
Scott, A. J. et al. International Cancer Microbiome Consortium consensus statement on the role of the human microbiome in carcinogenesis. Gut 68, 1624–1632 (2019).
Baxter, N. T., Koumpouras, C. C., Rogers, M. A., Ruffin, M. Tt & Schloss, P. D. DNA from fecal immunochemical test can replace stool for detection of colonic lesions using a microbiota-based model. Microbiome 4, 59 (2016).
Cammarota, G., Ianiro, G., Gasbarrini, A. & European, F. M. T. W. G. Faecal microbiota transplantation in clinical practice. Gut 67, 196–197 (2018).
Acknowledgements
J.Y. is supported by the Science and Technology Program Grant Shenzhen (JCYJ20170413161534162) and the Health and Medical Research Fund Hong Kong (17160862). Both authors are supported by grants from the CUHK Faculty of Medicine on Microbiota Research and the CUHK Vice-Chancellor’s Discretionary Fund.
Author information
Authors and Affiliations
Contributions
S.H.W. and J.Y. discussed the contents, wrote, reviewed and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wong, S.H., Yu, J. Gut microbiota in colorectal cancer: mechanisms of action and clinical applications. Nat Rev Gastroenterol Hepatol 16, 690–704 (2019). https://doi.org/10.1038/s41575-019-0209-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41575-019-0209-8
This article is cited by
-
The incidence of early onset colorectal cancer in Aotearoa New Zealand: 2000–2020
BMC Cancer (2024)
-
Analysis of differences in intestinal flora associated with different BMI status in colorectal cancer patients
Journal of Translational Medicine (2024)
-
Multi-omic profiling reveals associations between the gut microbiome, host genome and transcriptome in patients with colorectal cancer
Journal of Translational Medicine (2024)
-
Exosomes secreted by Fusobacterium nucleatum-infected colon cancer cells transmit resistance to oxaliplatin and 5-FU by delivering hsa_circ_0004085
Journal of Nanobiotechnology (2024)
-
Sex differences in colorectal cancer: with a focus on sex hormone–gut microbiome axis
Cell Communication and Signaling (2024)