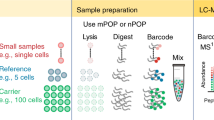
Abstract
Mass spectrometry (MS)-based proteomics typically employs multistep sample-preparation workflows that are subject to sample contamination and loss. We report an in-StageTip method for performing sample processing, from cell lysis through elution of purified peptides, in a single, enclosed volume. This robust and scalable method largely eliminates contamination or loss. Peptides can be eluted in several fractions or in one step for single-run proteome analysis. In one day, we obtained the largest proteome coverage to date for budding and fission yeast, and found that protein copy numbers in these cells were highly correlated (R2 = 0.78). Applying the in-StageTip method to quadruplicate measurements of a human cell line, we obtained copy-number estimates for 9,667 human proteins and observed excellent quantitative reproducibility between replicates (R2 = 0.97). The in-StageTip method is straightforward and generally applicable in biological or clinical applications.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Altelaar, A.M. & Heck, A.J. Trends in ultrasensitive proteomics. Curr. Opin. Chem. Biol. 16, 206–213 (2012).
de Godoy, L.M. et al. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature 455, 1251–1254 (2008).
Thakur, S.S. et al. Deep and highly sensitive proteome coverage by LC-MS/MS without prefractionation. Mol. Cell. Proteomics 10, M110 003699 (2011).
Kocher, T., Swart, R. & Mechtler, K. Ultra-high-pressure RPLC hyphenated to an LTQ-Orbitrap Velos reveals a linear relation between peak capacity and number of identified peptides. Anal. Chem. 83, 2699–2704 (2011).
Nagaraj, N. et al. System-wide perturbation analysis with nearly complete coverage of the yeast proteome by single-shot ultra HPLC runs on a bench top Orbitrap. Mol. Cell. Proteomics 11, M111 013722 (2012).
Yamana, R. et al. Rapid and deep profiling of human induced pluripotent stem cell proteome by one-shot NanoLC-MS/MS analysis with meter-scale monolithic silica columns. J. Proteome Res. 12, 214–221 (2013).
Michalski, A. et al. Mass spectrometry-based proteomics using Q Exactive, a high-performance benchtop quadrupole Orbitrap mass spectrometer. Mol. Cell. Proteomics 10, M111 011015 (2011).
Shevchenko, A., Wilm, M., Vorm, O. & Mann, M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858 (1996).
Chen, E.I., McClatchy, D., Park, S.K. & Yates, J.R. III. Comparisons of mass spectrometry compatible surfactants for global analysis of the mammalian brain proteome. Anal. Chem. 80, 8694–8701 (2008).
Nagaraj, N., Lu, A., Mann, M. & Wisniewski, J.R. Detergent-based but gel-free method allows identification of several hundred membrane proteins in single LC-MS runs. J. Proteome Res. 7, 5028–5032 (2008).
Manza, L.L., Stamer, S.L., Ham, A.J., Codreanu, S.G. & Liebler, D.C. Sample preparation and digestion for proteomic analyses using spin filters. Proteomics 5, 1742–1745 (2005).
Wisniewski, J.R., Zougman, A., Nagaraj, N. & Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362 (2009).
Ethier, M., Hou, W., Duewel, H.S. & Figeys, D. The proteomic reactor: a microfluidic device for processing minute amounts of protein prior to mass spectrometry analysis. J. Proteome Res. 5, 2754–2759 (2006).
Zhou, H., Ning, Z., Wang, F., Seebun, D. & Figeys, D. Proteomic reactors and their applications in biology. FEBS J. 278, 3796–3806 (2011).
Rappsilber, J., Mann, M. & Ishihama, Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 (2007).
Ong, S.E. et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 (2002).
Cox, J. & Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008).
Cox, J. & Mann, M. 1D and 2D annotation enrichment: a statistical method integrating quantitative proteomics with complementary high-throughput data. BMC Bioinformatics 13, S12 (2012).
Poulsen, J.W., Madsen, C.T., Young, C., Poulsen, F.M. & Nielsen, M.L. Using guanidine-hydrochloride for fast and efficient protein digestion and single-step affinity-purification mass spectrometry. J. Proteome Res. 12, 1020–1030 (2013).
Leon, I.R., Schwammle, V., Jensen, O.N. & Sprenger, R.R. Quantitative assessment of in-solution digestion efficiency identifies optimal protocols for unbiased protein analysis. Mol. Cell. Proteomics 12, 2992–3005 (2013).
Peng, M. et al. Protease bias in absolute protein quantitation. Nat. Methods 9, 524–525 (2012).
Wisniewski, J.R. et al. Extensive quantitative remodeling of the proteome between normal colon tissue and adenocarcinoma. Mol. Syst. Biol. 8, 611 (2012).
Picotti, P., Bodenmiller, B., Mueller, L.N., Domon, B. & Aebersold, R. Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell 138, 795–806 (2009).
Nieduszynski, C.A., Hiraga, S., Ak, P., Benham, C.J. & Donaldson, A.D. OriDB: a DNA replication origin database. Nucleic Acids Res. 35, D40–D46 (2007).
Ghaemmaghami, S. et al. Global analysis of protein expression in yeast. Nature 425, 737–741 (2003).
Gunaratne, J. et al. Extensive mass spectrometry-based analysis of the fission yeast proteome: The S. pombe PeptideAtlas. Mol. Cell. Proteomics 12, 1741–1751 (2013).
Marguerat, S. et al. Quantitative analysis of fission yeast transcriptomes and proteomes in proliferating and quiescent cells. Cell 151, 671–683 (2012).
Powell, S. et al. eggNOG v3.0: orthologous groups covering 1133 organisms at 41 different taxonomic ranges. Nucleic Acids Res. 40, D284–D289 (2012).
Zeiler, M., Straube, W.L., Lundberg, E., Uhlen, M. & Mann, M. A protein epitope signature Tag (PrEST) library allows SILAC-based absolute quantification and multiplexed determination of protein copy numbers in cell lines. Mol. Cell. Proteomics 11, O111.009613 (2012).
Nagaraj, N. et al. Deep proteome and transcriptome mapping of a human cancer cell line. Mol. Syst. Biol. 7, 548 (2011).
Beck, M. et al. The quantitative proteome of a human cell line. Mol. Syst. Biol. 7, 549 (2011).
Schwanhausser, B. et al. Global quantification of mammalian gene expression control. Nature 473, 337–342 (2011).
Cristobal, I. et al. PP2A impaired activity is a common event in acute myeloid leukemia and its activation by forskolin has a potent anti-leukemic effect. Leukemia 25, 606–614 (2011).
Kar, R., Singha, P.K., Venkatachalam, M.A. & Saikumar, P. A novel role for MAP1 LC3 in nonautophagic cytoplasmic vacuolation death of cancer cells. Oncogene 28, 2556–2568 (2009).
Tanida, I., Minematsu-Ikeguchi, N., Ueno, T. & Kominami, E. Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy. Autophagy 1, 84–91 (2005).
Forsburg, S.L. Eukaryotic MCM proteins: beyond replication initiation. Microbiol. Mol. Biol. Rev. 68, 109–131 (2004).
Williams, R.S., Williams, J.S. & Tainer, J.A. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem. Cell Biol. 85, 509–520 (2007).
Levin, J.Z. et al. Comprehensive comparative analysis of strand-specific RNA sequencing methods. Nat. Methods 7, 709–715 (2010).
Schaab, C., Geiger, T., Stoehr, G., Cox, J. & Mann, M. Analysis of high accuracy, quantitative proteomics data in the MaxQB database. Mol. Cell. Proteomics 11, M111 014068 (2012).
Masuda, T., Tomita, M. & Ishihama, Y. Phase transfer surfactant-aided trypsin digestion for membrane proteome analysis. J. Proteome Res. 7, 731–740 (2008).
Scheltema, R.A. & Mann, M. SprayQc: a real-time LC-MS/MS quality monitoring system to maximize uptime using off the shelf components. J. Proteome Res. (11 May 2012).
Cox, J. et al. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 (2011).
Acknowledgements
We thank S. Braun and P. Georgescu (Ludwig Maximilian University of Munich) for providing us with fission yeast pellets, F. Sacco, M. Steger, D. Walther and M. Räschle for help concerning biological interpretations. M. Hein, A. Dalfovo, C. Schaab and J. Cox helped with figures, videos and bioinformatics analysis of our data sets. Work in M.M.'s laboratory is supported by the Max Planck Society for the Advancement of Science and PROSPECT, a 7th framework program of the European Union (grant agreement HEALTH-F4-2008-201648).
Author information
Authors and Affiliations
Contributions
N.A.K. and M.M. developed and invented the method; G.P. and N.N. contributed in the developments; N.A.K., G.P., I.P. and N.N. performed the experiments; and N.A.K., G.P., N.N. and M.M. designed and interpreted the experiments, and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–11 and Supplementary Note (PDF 1672 kb)
Supplementary Table 1
Protein identifications and protein copy number estimations of S.cerevisiae, S.pombe and HeLa cells. (XLSX 1707 kb)
Supplementary Table 2
Orthologs of S.cerevisiae, S.pombe and HeLa cells and their protein copy number estimations. (XLSX 931 kb)
Supplementary Table 3
Buffer compositions and materials for the iST method. (XLSX 12 kb)
iST sample-preparation video tutorial.
The video tutorial shows all steps of the iST sample preparation workflow. It also shows how to troubleshoot the method, if necessary. (MP4 27589 kb)
Rights and permissions
About this article
Cite this article
Kulak, N., Pichler, G., Paron, I. et al. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods 11, 319–324 (2014). https://doi.org/10.1038/nmeth.2834
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.2834
This article is cited by
-
Proteomics of prostate cancer serum and plasma using low and high throughput approaches
Clinical Proteomics (2024)
-
One-Tip enables comprehensive proteome coverage in minimal cells and single zygotes
Nature Communications (2024)
-
FOXO1-mediated lipid metabolism maintains mammalian embryos in dormancy
Nature Cell Biology (2024)
-
The molecular basis of translation initiation and its regulation in eukaryotes
Nature Reviews Molecular Cell Biology (2024)
-
Biological big-data sources, problems of storage, computational issues, and applications: a comprehensive review
Knowledge and Information Systems (2024)