Abstract
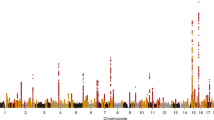
As end-stage renal disease (ESRD) has a four times higher incidence in African Americans compared to European Americans, we hypothesized that susceptibility alleles for ESRD have a higher frequency in the West African than the European gene pool. We carried out a genome-wide admixture scan in 1,372 ESRD cases and 806 controls and found a highly significant association between excess African ancestry and nondiabetic ESRD (lod score = 5.70) but not diabetic ESRD (lod = 0.47) on chromosome 22q12. Each copy of the European ancestral allele conferred a relative risk of 0.50 (95% CI = 0.39–0.63) compared to African ancestry. Multiple common SNPs (allele frequencies ranging from 0.2 to 0.6) in the gene encoding nonmuscle myosin heavy chain type II isoform A (MYH9) were associated with two to four times greater risk of nondiabetic ESRD and accounted for a large proportion of the excess risk of ESRD observed in African compared to European Americans.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
U.S. Renal Data System. USRDS 2007 Annual Data Report: Atlas of Chronic Kidney Disease and End-Stage Renal Disease in the United States (ed. National Institute of Diabetes and Digestive and Kidney Disease) (National Institute of Diabetes and Digestive and Kidney Disease, Bethesda, Maryland, 2007).
Klag, M.J. End-stage renal disease in African-American and white men - 16-year MRFIT findings. J. Am. Med. Assoc. 277, 1293–1298 (1997).
Tarver-Carr, M.E. et al. Excess risk of chronic kidney disease among African-American versus white subjects in the United States: A population-based study of potential explanatory factors. J. Am. Soc. Nephrol. 13, 2363–2370 (2002).
Perneger, T.V., Whelton, P.K. & Klag, M.J. Race and end-stage renal disease. Socioeconomic status and access to health care as mediating factors. Arch. Intern. Med. 155, 1201–1208 (1995).
Lei, H.H., Perneger, T.V., Klag, M.J., Whelton, P.K. & Coresh, J. Familial aggregation of renal disease in a population-based case-control study. J. Am. Soc. Nephrol. 9, 1270–1276 (1998).
Freedman, B.I., Spray, B.J., Tuttle, A.B. & Buckalew, V.M. The familial risk of end-stage renal disease in African Americans. Am. J. Kidney Dis. 21, 387–393 (1993).
Fox, C.S. et al. Genomewide linkage analysis to serum creatinine, GFR, and creatinine clearance in a community-based population: the Framingham Heart Study. J. Am. Soc. Nephrol. 15, 2457–2461 (2004).
Hunt, S.C. et al. Linkage of serum creatinine and glomerular filtration rate to chromosome 2 in Utah pedigrees. Am. J. Hypertens. 17, 511–515 (2004).
Chakraborty, R. & Weiss, K.M. Admixture as a tool for finding linked genes and detecting that difference from allelic association between loci. Proc. Natl. Acad. Sci. USA 85, 9119–9123 (1988).
Stephens, J.C., Briscoe, D. & O'Brien, S.J. Mapping by admixture linkage disequilibrium in human populations: limits and guidelines. Am. J. Hum. Genet. 55, 809–824 (1994).
McKeigue, P.M. Mapping genes that underlie ethnic differences in disease risk: methods for detecting linkage in admixed populations, by conditioning on parental admixture. Am. J. Hum. Genet. 63, 241–251 (1998).
Smith, M.W. & O'Brien, S.J. Mapping by admixture linkage disequilibrium: advances, limitations and guidelines. Nat. Rev. Genet. 6, 623–632 (2005).
Smith, M.W. et al. A high-density admixture map for disease gene discovery in African Americans. Am. J. Hum. Genet. 74, 1001–1013 (2004).
Patterson, N. et al. Methods for high-density admixture mapping of disease genes. Am. J. Hum. Genet. 74, 979–1000 (2004).
McKeigue, P.M., Carpenter, J.R., Parra, E.J. & Shriver, M.D. Estimation of admixture and detection of linkage in admixed populations by a Bayesian approach: application to African-American populations. Ann. Hum. Genet. 64, 171–186 (2000).
Haiman, C.A. et al. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat. Genet. 39, 638–644 (2007).
Reich, D. et al. A whole-genome admixture scan finds a candidate locus for multiple sclerosis susceptibility. Nat. Genet. 37, 1113–1118 (2005).
Iyengar, S.K. et al. Linkage analysis of candidate loci for end-stage renal disease due to diabetic nephropathy. J. Am. Soc. Nephrol. 14, S195–S201 (2003).
Remuzzi, G., Benigni, A. & Remuzzi, A. Mechanisms of progression and regression of renal lesions of chronic nephropathies and diabetes. J. Clin. Invest. 116, 288–296 (2006).
Su, A.I. et al. Large-scale analysis of the human and mouse transcriptomes. Proc. Natl. Acad. Sci. USA 99, 4465–4470 (2002).
Kunishima, S., Matsushita, T., Hamaguchi, M. & Saito, H. Identification and characterization of the first large deletion of the MYH9 gene associated with MYH9 disorders. Eur. J. Haematol. 80, 540–544 (2008).
Kunishima, S. et al. Mapping of a gene for May-Hegglin anomaly to chromosome 22q. Hum. Genet. 105, 379–383 (1999).
Saito, H. & Kunishima, S. Historical hematology: May-Hegglin anomaly. Am. J. Hematol. 83, 304–306 (2008).
Kelley, M.J., Jawien, W., Ortel, T.L. & Korczak, J.F. Mutation of MYH9, encoding non-muscle myosin heavy chain A, in May-Hegglin anomaly. Nat. Genet. 26, 106–108 (2000).
Seri, M. et al. Epstein syndrome: another renal disorder with mutations in the nonmuscle myosin heavy chain 9 gene. Hum. Genet. 110, 182–186 (2002).
Kopp, J.B. et al. MYH9 is a major-effect risk gene for focal segmental glomerulosclerosis. Nat. Genet. advance online publication, doi:10.1038/ng.226 (14 September 2008).
D'Apolito, M., Guarnieri, V., Boncristiano, M., Zelante, L. & Savoia, A. Cloning of the murine non-muscle myosin heavy chain IIA gene ortholog of human MYH9 responsible for May-Hegglin, Sebastian, Fechtner, and Epstein syndromes. Gene 286, 215–222 (2002).
Marini, M. et al. Non-muscle myosin heavy chain IIA and IIB interact and co-localize in living cells: relevance for MYH9-related disease. Int. J. Mol. Med. 17, 729–736 (2006).
Wright, J.T. Jr et al. Effect of blood pressure lowering and antihypertensive drug class on progression of hypertensive kidney disease: results from the AASK trial. J. Am. Med. Assoc. 288, 2421–2431 (2002).
Knowler, W.C. et al. The Family Investigation of Nephropathy and Diabetes (FIND); design and methods. J. Diabetes Complications 19, 1–9 (2005).
Powe, N.R. et al. Choices for healthy outcomes in caring for end stage renal disease. Semin. Dial. 9, 9–11 (1996).
Nalls, M.A. et al. Admixture mapping of white cell count: genetic locus responsible for lower white blood cell count in the Health ABC and Jackson Heart studies. Am. J. Hum. Genet. 82, 81–87 (2008).
Hinds, D.A. et al. Whole-genome patterns of common DNA variation in three human populations. Science 307, 1072–1079 (2005).
The International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature 449, 851–861 (2007).
Kong, X. et al. A combined linkage-physical map of the human genome. Am. J. Hum. Genet. 75, 1143–1148 (2004).
Acknowledgements
We thank the study participants, staff, laboratories and physicians who participated in the FIND and the CHOICE Study at Dialysis Clinic, Inc. and at Johns Hopkins University. This study was also supported by the following research grants: U01DK070657, U01DK57304, K01DK067207 (W.H.L.K.), U01DK57292, and DK07024 (the CHOICE study) from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) and, in part, by the Intramural Research Program of the NIDDK. The CHOICE study was also supported in part by HS08365 from the Agency for Healthcare Research and Quality, Rockville, Maryland and HL62985 from the National Heart Lung and Blood Institute, Bethesda, Maryland. This work was supported by the National Center for Research Resources for the General Clinical Research Center (GCRC) grants: Case Western Reserve University M01-RR-000080; Wake Forest University M01-RR-07122; Harbor-UCLA Medical Center M01-RR-00425; College of Medicine - University of California Irvine M01-RR-00827-29; Frederic C. Bartter M01-RR-01346. D.R. was supported by a Burroughs Wellcome Career Development Award in the Biomedical Sciences.
This research was supported in part by the Intramural Research Program of the US National Institutes of Health, National Cancer Institute, Center for Cancer Research. This project has been funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract N01-CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does it mention of trade names, commercial products, or organizations imply endorsement by the US government.
Author information
Authors and Affiliations
Consortia
Corresponding author
Additional information
A full list of members is provided in the Supplementary Note.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1–7, Supplementary Note (PDF 627 kb)
Rights and permissions
About this article
Cite this article
Kao, W., Klag, M., Meoni, L. et al. MYH9 is associated with nondiabetic end-stage renal disease in African Americans. Nat Genet 40, 1185–1192 (2008). https://doi.org/10.1038/ng.232
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.232
This article is cited by
-
Integrated proteome and malonylome analyses reveal the potential meaning of TLN1 and ACTB in end-stage renal disease
Proteome Science (2023)
-
The evolving story of apolipoprotein L1 nephropathy: the end of the beginning
Nature Reviews Nephrology (2022)
-
Undercutting efforts of precision medicine: roadblocks to minority representation in breast cancer clinical trials
Breast Cancer Research and Treatment (2021)
-
A focus on the association of Apol1 with kidney disease in children
Pediatric Nephrology (2021)
-
Association of MYH9-rs3752462 polymorphisms with chronic kidney disease among clinically diagnosed hypertensive patients: a case-control study in a Ghanaian population
Clinical Hypertension (2020)