Abstract
Myotonic dystrophy (DM) is the most common adult-onset muscular dystrophy with an estimated prevalence of 1/8000. There are two genetically distinct types, DM1 and DM2. DM2 is generally milder with more phenotypic variability than the classic DM1. Our previous data on co-segregation of heterozygous recessive CLCN1 mutations in DM2 patients indicated a higher than expected DM2 prevalence. The aim of this study was to determine the DM2 and DM1 frequency in the general population, and to explore whether the DM2 mutation functions as a modifier in other neuromuscular diseases (NMD) to account for unexplained phenotypic variability. We genotyped 5535 Finnish individuals: 4532 normal blood donors, 606 patients with various non-myotonic NMD, 221 tibial muscular dystrophy patients and their 176 healthy relatives for the DM2 and DM1 mutations. We also genotyped an Italian idiopathic non-myotonic proximal myopathy cohort (n=93) for the DM2 mutation. In 5496 samples analyzed for DM2, we found three DM2 mutations and two premutations. In 5511 samples analyzed for DM1, we found two DM1 mutations and two premutations. In the Italian cohort, we identified one patient with a DM2 mutation. We conclude that the DM2 mutation frequency is significantly higher in the general population (1/1830; P-value=0.0326) than previously estimated. The identification of DM2 mutations in NMD patients with clinical phenotypes not previously associated with DM2 is of particular interest and is in accord with the high overall prevalence. On the basis of our results, DM2 appears more frequent than DM1, with most DM2 patients currently undiagnosed with symptoms frequently occurring in the elderly population.
Similar content being viewed by others
INTRODUCTION
Myotonic dystrophy is an autosomal dominant multi-systemic neuromuscular disorder. Two genetically distinct diseases with clinical similarities but also distinct differences have been identified. Myotonic dystrophy type 1 (DM1, Steinert's disease, MIM #160900) is caused by a (CTG)n expansion mutation in the 3′ UTR of dystrophia myotonica protein kinase (DMPK) in chromosome 19q13.3;1, 2, 3 DM type 2 (DM2, MIM #602668) is caused by a (CCTG)n expansion mutation in the first intron of zinc-finger protein 9 (ZNF9) in chromosome 3q21.4, 5 Similar clinical features of DM1 and DM2 include myotonia and limb muscle weakness, pronounced distal in DM1 and proximal in DM2, and multi-organ involvement including cataracts, insulin resistance, elevated liver enzyme levels, male hypogonadism and cardiac conduction defects. Muscle atrophy, facial weakness, ptosis and frontal baldness are very prevalent in DM1, whereas muscle pain and hypertrophy of calf muscles are more characteristic for DM2. Serum CK levels are usually elevated. Overall, clinical manifestations in DM2 appear to be more variable and generally milder than those in classic adult-onset DM1 (for review see refs 6 and7).
In DM1 but not in DM2, the length of the expansion mutation generally correlates with the severity of the disease. Normal individuals have 5–37 (CTG)n repeats at the DMPK locus and the number of repeats in DM1 patients varies from mild late-onset cases with ≥50 repeats to the most severe congenital form of DM1 containing up to 4000 (CTG)n repeats.1 The four main categories of DM1 based on clinical outcome are (1) congenital, (2) childhood-onset, (3) classic adult-onset, and (4) late-onset/oligo-symptomatic. The distinction is not absolute but rather a continuum generally correlating with increasing length of the (CTG)n expansion, which tends to increase through successive generations causing genetic anticipation.
The DM2 expansion mutation in intron 1 of ZNF9 consists of several normally polymorphic elements first described as (TG)n(TCTG)n(CCTG)n, but variations with short intervening cryptic repeats (TCTG and/or GCTG) disrupting the (CCTG)n tract have also been reported.4, 5, 8, 9 It has been proposed that only uninterrupted (CCTG)n repeats are associated with the DM2 phenotype.5, 8, 9 The range of (CCTG)n repeats in patients varies between 55 and 11 000, but, unlike DM1, the threshold defining the difference between normal and disease-causing repeats has not yet been firmly established.5, 9 For DM2, correlation between repeat length and disease severity or age-of-onset has not been demonstrated. Phenotypic variability and the large size of most mutant repeats make this determination more complicated.
Before the identification of their respective genetic mutations, the combined prevalence of DM1 and DM2 has generally been estimated at 1 in 8000 (12.5 per 100 000), based on clinical ascertainment of patients. As this estimate is likely based mostly on more severely affected DM1 patients, DM1 is considered to be the most frequent muscular dystrophy in adults.10 However, in different populations prevalence estimates vary widely: In an Italian population from the regions around Padova and Northwest Tuscany, a prevalence of 9.31 per 100 000 inhabitants was reported,11 whereas a slightly higher prevalence (11.95 per 100 000) was reported in Northern Ireland,12 and lower in Belgrade (Serbia) (5.3 per 100 000).13 In Jewish communities, an average prevalence of 15.7 per 100 000 was reported in a study including 416 DM1 patients of which 307 were diagnosed genetically and 109 were clinically diagnosed.14 The same study reported even higher prevalences in two Jewish subgroups: 20.0 per 100 000 in Sephardim/Oriental Jews and 47.3 per 100 000 in Yemenite Jews. Among Jews, Ashkenazi had the lowest prevalence with 5.7 per 100 000. A relatively high prevalence of DM1 in the smaller Istria region in Croatia has been reported (18.1 per 100 000) based on clinical ascertainment and genetic confirmation.15 The highest known prevalence has been reported in the French Canadian population (1 in 475) because of a founder effect.16 In non-European populations much lower prevalence rates have been observed, for example a prevalence of 0.46 per 100 000 in Taiwan has been estimated based on DM1 patients and families identified by genetic analysis during 1990–2001 (96 subjects belonging to 26 families).17, 18
For DM2, there are currently no established prevalence estimates. DM2 is generally thought to be rarer than DM1, but large-scale population studies to confirm this have not been carried out. In Finland, 56 new DM2 patients were genetically diagnosed in 2004–2005, and a prevalence estimate of 1/10 000 based on molecular genetic testing has been reported for the Central Finland hospital district.19 However, new clinical ascertainment data suggest at least a twofold higher prevalence in Central Finland (unpublished data). In Germany, 267 mutation-verified DM2 molecular diagnoses were made between 2003 and 2005 compared with 277 DM1 diagnoses within the same period.19 These data support the notion that DM2 is more frequent than previously thought, possibly even as frequent as DM1. Because of a shared ancestral founder haplotype common to all Caucasian DM2 patients,4 there is no a priori reason to expect the frequency of DM2 mutation to be highly variable in different European populations.
Because of the wide spectrum of milder phenotypic presentations in DM2, the prevalence of the disease is not easy to estimate clinically. Tissue-related differences in repeat instability, modifier genes or other mechanisms may influence the length of (CCTG)n repeats needed to elicit symptoms in a given patient and within different organs. In DM2, there is no known correlation between repeat size and disease severity. Our experience clearly indicates a higher prevalence of very late onset and mild manifestations of DM2 than previously reported.20 Because the first descriptions of the clinical phenotype were based on families collected for linkage studies, there might have been an intrinsic ascertainment bias for more severe disease manifestations, resulting in a lack of very mild cases in the phenotypic spectrum. Our objective in this study was to determine the overall frequency of the DM2 and DM1 mutations in the general population.
SUBJECTS AND METHODS
Subjects
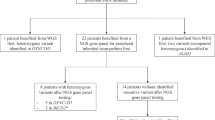
To study the frequency of DM2 and DM1 expansion mutations, we analyzed 4532 Finnish population control DNA samples obtained from anonymous blood donors. The Finnish population is a historically isolated population with very specific features regarding accumulation of certain recessive disorders based on limited numbers of founder populations and bottlenecks. For dominant disorders, the outcome is somewhat different and the Finnish population is expected to be a relatively representative Caucasian population in regard to the DM2 mutation, because of the ancestral founder haplotype existing in all Europe. The material was selected because of the availability of suitable and high-quality samples. To study the frequency of the DM mutations and to explore whether the presence of the DM2 (or DM1) mutation might function as a modifier in other neuromuscular diseases (NMD), we also studied an additional 1003 Finnish samples (862 independent chromosomes) consisting of 606 patients with various non-myotonic NMD, 221 tibial muscular dystrophy (TMD) patients, and 176 healthy relatives of the TMD patients from previous linkage studies. We also studied a cohort of 93 Italian patients with undetermined non-myotonic proximal myopathy or asymptomatic hyperCKemia, which were genotyped for the DM2 mutation only because of their proximal muscle phenotype. The study was approved by the IRB of Tampere University Hospital.
Methods
DNA isolated from peripheral blood leukocytes was genotyped for both the DM2 and DM1 mutations as previously described.4, 21, 22, 23 Briefly, first DM2 and DM1 repeats were amplified by PCR across the repeat under conditions in which only normal-sized alleles are amplified. Samples with a single allele (either two normal alleles of identical size, or only one normal allele because of resistance of expanded mutant alleles to PCR) were further studied using repeat-primed PCR (RP-PCR). The results for both the steps were obtained using fluorescent fragment analysis by capillary electrophoresis (ABI 3100 or ABI 3130 Genetic Analyzer) (Applied Biosystems, Foster City, CA, USA) and analyzed using Genotyper or GeneMapper software (Applied Biosystems). For the two DM2 premutation alleles from the NMD cohort, single genome equivalent amplification (small-pool PCR) was performed as previously described to determine the stability of the repeat region,9, 24 and sequencing of the repeat region was carried out to characterize the repeat number and interruptions of the (CCTG)n repeat in ZNF9, as previously described.9 A single-nucleotide polymorphism (SNP, rs1871922) in linkage disequilibrium (LD) with the DM2 repeat expansion mutation was also genotyped as previously described4, 9 for the samples with large, possibly unstable DM2 alleles in ZNF9.
Statistical analyses
To determine the statistical significance in the general population cohort, we modeled the presence of a DM mutation (either DM1 or DM2) as a binomial random variable, where in 4510 tries, we observed four mutation-positive cases. Using a likelihood function for the detection rate, P-value, we computed both an equal-tailed (ET) 95% interval estimate (excluding the bottom and top 2.5% of all values) and a maximum likelihood (ML) 95% interval estimate (all values in the interval have a likelihood above a threshold value). Computation of these estimates used the fact that the likelihood function is proportional to a beta(4+14 506+1) distribution. We then determined whether 1/8000 falls in either of the interval, and computed the chance that ≥4 mutations would be seen in 4510 trials if this were in fact the true mutation rate.
Statistical significance in the combined general population and NMD cohorts was modeled similarly using a likelihood function that was proportional to a beta(5+15 495+1) distribution, where in 5500 tries, we observed five mutation-positive cases. We again determined whether 1/8000 falls in either interval, and computed the chance that ≥5 mutations would be seen in 5500 trials if this were in fact the true mutation rate.
Similarly, to determine statistical significance for the DM2 mutation alone in the general population cohort, we modeled the mutation frequency using a likelihood function that was proportional to a beta(3+15 497+1) distribution, where in 5500 trials, we observed three mutation-positive cases. We then determined whether 1/8000 falls in either interval, and computed the chance that ≥3 mutations would be seen in 5500 trials if this were in fact the true mutation rate.
RESULTS
General population cohort
The population cohort consisted of 4532 anonymous blood donor DNA samples. Results for allele sizes were obtained from 4508 samples for DM2 and from 4520 for DM1 giving very low failure rates (0.53% for DM2 and 0.26% for DM1). A single allele was seen in 12.6% (572) of samples at the ZNF9 locus and in 20.7% (938) of samples at the DMPK locus. Using RP-PCR on these single-allele samples, two DM2 and two DM1 mutation-positive samples were identified (Table 1). One of the DM1 mutations had a (CTG)80 repeat allele, whereas for the other, the exact repeat number of the expanded allele could not be determined. For the two DM2 mutations, the exact (CCTG)n repeat number could not be determined with the methods used. Both DM2 mutation-positive samples showed homozygosity for the C allele of SNP rs1871922 in LD with the DM2 expansion mutation.
In addition to the normal and mutant alleles, a small number of enlarged alleles that fall in the range between normal and mutant were identified. Such enlarged alleles were found in ZNF9 in nine samples, of which five showed a pattern consistent with an unstable (CCTG)n repeat on RP-PCR. These nine samples were genotyped for SNP rs1871922 and the majority (n=6) were homozygous (C/C) and three samples were heterozygous (A/C).
In DMPK, one premutation allele of approximately (CTG)45 repeats was identified and in 13 other samples, alleles in the range of 35–40 repeats were identified by PCR across the repeat region. In seven additional samples, RP-PCR suggested an enlarged allele of unknown repeat size.
NMD patient cohort
Among 988 samples with available molecular diagnostic DM2 data, one DM2 mutation was found (Table 1; Figure 1). Approximately 11% of the individuals studied showed one single allele for the repeat tract in intron 1 of ZNF9. Possible large alleles were detected in six individuals by RP-PCR. Sequencing of these DM2 repeat alleles revealed two samples with uninterrupted short repeat expansions of (CCTG)24 and (CCTG)25 repeats. These expanded alleles were also found to be unstable in the small-pool PCR, and the samples were heterozygous for SNP rs1871922 (A/C).9 The patient with a DM2 mutation was previously diagnosed with genetically verified TMD, but with unusually marked proximal muscle involvement. Clinically the patient had no myotonia. EMG studies were not performed, and the patient is no longer available for follow-up studies. Of the two patients with uninterrupted unstable (CCTG)24−25 tracts, one was diagnosed with a mitochondrial DNA 3243A>G mutation causing Mitochondrial myopathy, Encephalopathy, Lactic Acidosis and Stroke-like episodes (MELAS) syndrome;25, 26 the other patient had an undetermined asymmetric muscle disorder with pseudohypertrophy of one leg and mild atrophy of the other leg. In light of the lack of any DM-like phenotype, the co-segregating uninterrupted unstable (CCTG)24−25 tract did not appear to have any discernable effect on the phenotype in these two patients.
Representative results for DM2 mutation genotyping by PCR and RP-PCR. PCR across the (CCTG)DM2 in intron 1 of ZNF9 showed either a single amplifiable allele (a) or two amplifiable alleles (b), which excludes DM2 mutation. A patient with DM2 mutation from the population cohort showed a peak pattern in ZNF9 RP-PCR characteristic for the DM2 (CCTG)n repeat expansion mutation (c and d). In the zoomed-in figure (d) adjacent stutter peaks can be seen with base pair difference of 4 bp. Enlarged unstable (but not actually expanded) DM2 alleles have a distinct pattern in RP-PCR (e and f). In the zoomed-in figure (f), the stutter peaks can be seen, but the pattern ends rapidly. Figures (g) and (h) show a typical example of a DM2 negative sample in RP-PCR. Stutter peaks characteristic to DM2 mutation are not seen in the zoomed-in figure (h).
Among 991 samples with available molecular diagnostic DM1 data, there were no DM1 mutations (Table 1). One borderline pre-mutation allele of 37 repeats was found in one individual. Approximately 19% of samples were homozygous for the DM1 repeat in DMPK showing one allele size in the locus.
In conclusion, combining our general population cohort and the NMD cohort, we found three DM2 mutations in 5496 individuals and two DM1 mutations in 5511 individuals. On the basis of these results, the frequency of DM mutations is approximately 1 in 1830 for DM2 and 1 in 2760 for DM1. Because all patients with myotonia were excluded from the NMD cohort and DM mutations segregate independently from the underlying cause of disease in these patients, this cohort can be considered as an unselected population with respect to both DM mutations.
Cohort of proximal myopathy or asymptomatic hyperCKemia
A cohort consisting of 93 Italian patients with idiopathic proximal myopathy or asymptomatic hyperCKemia was analyzed for the DM2 mutation by PCR across the DM2 repeat and RP-PCR. In this cohort, one DM2 mutation-positive patient was found (Table 1). This patient showed waddling gait, proximal weakness and Gower's sign at the age of 49 years, whereas CK levels were normal. EMG showed findings consistent with a myopathic pattern without myotonic discharges.
Statistical analyses
The previously reported prevalence for DM is 1 in 8000 (0.000125=1.25E−4).10 Using a likelihood function to model the presence of a DM mutation (either DM1 or DM2) as a binomial random variable in 4510 anonymous blood donor samples with available genotypes (general population cohort), the observed frequency of four mutation-positive carriers was significantly higher than expected by chance (point estimate (PE): 8.9E−4, ET 95% interval (3.6E−4, 2.27E−3), ML 95% interval (2.7E−4, 2.09E−3), P-value=0.0027). Similarly, the observed frequency of five mutation-positive carriers in the combined general population and NMD cohorts (n=5500) was significantly higher than expected by chance (PE: 9.1E−4, ET: (4.0E−4, 2.12E−3), ML: (3.2E−4, 1.98E−3), P-value=0.0007). The frequency of the DM2 mutation by itself was also significantly more frequent (PE: 5.45E−4, ET: (1.98E−4, 1.59E−3), ML: (1.30E−4, 1.44E−3), P=0.0326 for the general population and NMD cohorts combined). These results are presented in Table 2. Starting with the null hypothesis that the true rate of the DM prevalence is 1/8000 and testing this by measuring the status of 5500 individuals with a required type I error α≤0.05, observing ≥3 mutation-positive DM cases leads us to reject the null hypothesis. If the true rate is 5/5500 (or 1/1100), we have 87.5% power to correctly identify a difference. Thus, our data clearly indicate that DM (DM1 and DM2 combined) and DM2 alone are significantly more frequent than could be expected for the population based on previous estimates.
DISCUSSION
Early estimates of disease frequency for DM relied on clinical ascertainment of patients and have resulted in a calculated overall prevalence of 1/8000.10 As the advent of molecular genetic testing for DM1, several prevalence studies have been reported,11, 12, 13, 14, 15, 17 but so far no large-scale population studies have been performed. Most of the reported studies were based on the number of diagnosed patients related to the corresponding total population in a certain region. In regions with good diagnostic services, this type of ascertainment may be close to 100% for the subcategories congenital and adult-onset DM1. However, the actual DM1 mutation frequency may be higher because of the known existence of a large group of clinically undiagnosed oligosymptomatic mild forms and some undiagnosed childhood-onset forms. For DM2, clinically based ascertainment of patients is even more difficult because of the large phenotypic variability and a large number of individuals with milder symptoms who remain undiagnosed. Since the availability of DM2 molecular diagnostics, our experience indicated that DM2 is far more common than previously estimated.27, 28 Milder phenotypes with prominent myalgia may easily be misdiagnosed as fibromyalgia,27 and patients with onset of slowly progressive proximal muscle weakness after age 70 years may not be referred for neuromuscular investigations. Further evidence for a large proportion of undiagnosed DM2 patients came from our study on diagnosed DM2 patients showing a disproportionally high number of co-segregating heterozygous recessive CLCN1 mutations.28 This study directly suggested that DM2 patients with co-segregating CLCN1 could be more easily identified and diagnosed than DM2 patients without the modifier allele, and consequently that the majority of DM2 patients remains undiagnosed even in clinical centers with considerable experience with DM2.
Our present study investigated the prevalence of the DM2 and DM1 mutations in a cohort of 5535 individuals consisting of anonymous blood donors and patients with various determined and undetermined neuromuscular disorders with exclusion of any myotonic symptoms. The finding of three DM2 mutations in this cohort suggests a mutation frequency of 1 in 1830, which is >fourfold higher than any previous estimates. In the same cohort we identified two DM1 mutations, suggesting a mutation frequency of 1 in 2760 in the general population. Compared with previous prevalence estimates for DM, this is almost threefold higher. These results are significantly different to the previous DM prevalence estimate of 1/8000 (4/4510, P-value=0.0027 for the general population cohort, and 5/5500, P-value=0.0007 when general and NMD cohorts are combined).
The higher than expected DM1 mutation frequency is probably because of a large number of smaller expansion alleles, which are known to be asymptomatic or cause very mild symptoms that can be confused with normal aging in elderly individuals.29 Clinically this may indicate that a number of elderly patients with cataracts or ptosis may, in fact, have DM1 as the underlying cause. However, if these patients do not develop other symptoms indicative of DM1 during their lifetime there is no need to change clinical practice.
However, with DM2 the situation is different, because all DM2 mutation carriers are expected to develop disease symptoms, and patients currently remain undiagnosed and their symptoms are currently incompletely understood. This may cause significant differences in DM2 ascertainment in different populations, because the prevailing practice for directing a patient for genetic testing of DM2 mutation may vary. In the most typical families selected for linkage studies, the DM2 mutation was 100% penetrant. However, it is not settled if this is true in all situations or if the phenotype can extend beyond the currently known variation. As the DM2 mutation may also cause cardiac conduction defects and sudden cardiac death as early as in middle age,30 the timely identification of DM2 mutations in carriers is of major clinical importance. Mutation carriers may present to a wide range of clinical specialties: neurology, cardiology, internal medicine, ophthalmology, rheumatology, endocrinology and so on. The mutation frequency determined in this study suggests a need for increased consideration of DM2 as a possible cause of symptoms. At least those patients undergoing neurological evaluation should be assessed by molecular diagnostic testing with a rather low phenotypic threshold.8
Because the DM2 mutation has a single European founder haplotype,4 the frequency of DM2 in the Finnish population is suggestive for a high-mutation frequency also in other Caucasian populations. However, specific historical population bottlenecks and genetic drift may cause somewhat skewed frequencies in different sub-populations. Samples from healthy blood donors are expected to be relatively representative of the population at large from which they are drawn.
Uninterrupted unstable DM2 repeat tracts of (CCTG)24 and (CCTG)25 repeats were found in two individuals from the NMD cohort. One of these patients had a diagnosis of molecularly verified MELAS syndrome, whereas the other had an undetermined asymmetric muscle hypertrophy in one leg and atrophy in the other. However, deleterious consequences of these small expansion alleles on the phenotype in these individuals are unlikely. In the general population cohort we found nine samples with possibly unstable enlarged alleles, suggesting a DM2 premutation. These nine samples in addition to the two small expansion samples found in the NMD cohort were all either heterozygous or homozygous for the C allele at SNP rs1871922, which is in LD with the DM2 haplotype.4 The occurrence of such uninterrupted unstable repeat tracts is compatible with the recent identification of DM2 premutation alleles.9
The single DM2 mutation in the NMD cohort was found in a patient with a genetically confirmed diagnosis of TMD. Besides the conventional weakness of ankle dorsiflexion, this patient had an unusually marked proximal muscle involvement at the age of 55 years. The DM2 mutation mainly affects proximal muscles, suggesting that the marked proximal muscle involvement in this particular DM2-positive TMD patient could be associated with the co-segregating DM2 mutation, although its impact cannot be definitively determined. Most TMD patients have a highly selective involvement of the anterior compartment muscles of the lower legs and marked proximal lower limb weakness occurring only after the age of 70–75 years. In a study of 209 TMD patients, 9% had very unusual phenotypes, including onset in proximal muscles, despite an identical TTN founder mutation in all patients.31 The reason for these phenotypic variations remains unknown and was not associated with a co-segregating DM2 mutation. In the Italian cohort with 93 patients of undetermined non-myotonic proximal myopathy or asymptomatic hyperCKemia, one patient proved to have a molecular diagnosis of DM2. The fact that myotonia is frequently absent in DM2 patients, even on EMG examination, is not a new observation.7, 19 However, the identification of DM2 in a patient without otherwise suggestive features of the disease is of interest and suggests that the DM2 diagnosis may easily be missed in a neuromuscular diagnostic setting because of incomplete or uncharacteristic phenotypic expression.
Two DM1 mutations were identified in the general population cohort. A (CTG)80 allele was identified for one individual, whereas for the other, size could not be determined. Two premutation alleles were found in both the population cohort ((CTG)45) and the NMD cohort ((CTG)37).
Taken together our data indicate that mutations for DM (DM1 and DM2) are much more prevalent than previously estimated, and that DM2 may even be the most commonly inherited muscle disease in the European populations. The results also indicate that the vast majority of DM2 patients currently remain undiagnosed. At least two conclusions can be drawn: first, DM2 patients with symptoms for myalgia, muscle weakness in advanced age, insulin resistance or cardiac conduction abnormalities are currently not correctly identified, and second, a more comprehensive study of the natural history to characterize the entire spectrum of disease presentations is urgently needed for DM2.
References
Brook JD, McCurrach ME, Harley HG et al: Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell 1992; 69: 385.
Fu YH, Pizzuti A, Fenwick Jr RG et al: An unstable triplet repeat in a gene related to myotonic muscular dystrophy. Science 1992; 255: 1256–1258.
Mahadevan M, Tsilfidis C, Sabourin L et al: Myotonic dystrophy mutation: an unstable CTG repeat in the 3′ untranslated region of the gene. Science 1992; 255: 1253–1255.
Bachinski LL, Udd B, Meola G et al: Confirmation of the type 2 myotonic dystrophy (CCTG)n expansion mutation in patients with proximal myotonic myopathy/proximal myotonic dystrophy of different European origins: a single shared haplotype indicates an ancestral founder effect. Am J Hum Genet 2003; 73: 835–848.
Liquori CL, Ricker K, Moseley ML et al: Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science 2001; 293: 864–867.
Machuca-Tzili L, Brook D, Hilton-Jones D : Clinical and molecular aspects of the myotonic dystrophies: a review. Muscle Nerve. 2005; 32: 1–18.
Krahe R, Bachinski LL, Udd B : Myotonic dystrophy type 2: clinical and genetic aspects; in Ashizawa T, Wells D (eds): Genetic Instabilities and Neurological Diseases. USA: Academic Press, 2006, 2nd edn, pp 131–150.
Liquori CL, Ikeda Y, Weatherspoon M et al: Myotonic dystrophy type 2: human founder haplotype and evolutionary conservation of the repeat tract. Am J Hum Genet 2003; 73: 849–862.
Bachinski LL, Czernuszewicz T, Ramagli LS et al: Premutation allele pool in myotonic dystrophy type 2. Neurology 2009; 72: 490–497.
Harper PS : Myotonic Dystrophy, 3rd edn. London, UK: W.B. Saunders, 2001.
Siciliano G, Manca M, Gennarelli M et al: Epidemiology of myotonic dystrophy in Italy: re-apprisal after genetic diagnosis. Clin Genet 2001; 59: 344–349.
Magee A, Nevin NC : The epidemiology of myotonic dystrophy in Northern Ireland. Community Genet 1999; 2: 179–183.
Mladenovic J, Pekmezovic T, Todorovic S et al: Epidemiology of myotonic dystrophy type 1 (Steinert disease) in Belgrade (Serbia). Clin Neurol Neurosurg 2006; 108: 757–760.
Segel R, Silverstein S, Lerer I et al: Prevalence of myotonic dystrophy in Israeli Jewish communities: inter-community variation and founder premutations. Am J Med Genet A 2003; 119: 273–278.
Medica I, Markovic D, Peterlin B : Genetic epidemiology of myotonic dystrophy in Istria, Croatia. Acta Neurol Scand 1997; 95: 164–166.
Bouchard G, Roy R, Declos M, Mathieu J, Kouladjian K : Origin and diffusion of the myotonic dystrophy gene in the Saguenay region (Quebec). Can J Neurol Sci 1989; 16: 119–122.
Hsiao KM, Chen SS, Li SY et al: Epidemiological and genetic studies of myotonic dystrophy type 1 in Taiwan. Neuroepidemiology 2003; 22: 283–289.
Ashizawa T, Epstein HF : Ethnic distribution of myotonic dystrophy gene. Lancet 1991; 338: 642–643.
Udd B, Meola G, Krahe R et al: 140th ENMC International Workshop: myotonic dystrophy DM2/PROMM and other myotonic dystrophies with guidelines on management. Neuromuscul Disord 2006; 16: 403–413.
Day JW, Ricker K, Jacobsen JF et al: Myotonic dystrophy type 2: molecular, diagnostic and clinical spectrum. Neurology 2003; 60: 657–664.
Sallinen R, Vihola A, Bachinski LL et al: New methods for molecular diagnosis and demonstration of the (CCTG)n mutation in myotonic dystrophy type 2 (DM2). Neuromuscul Disord 2004; 14: 274–283.
Sermon K, Seneca S, De Rycke M et al: PGD in the lab for triplet repeat diseases – myotonic dystrophy, Huntington's disease and Fragile-X syndrome. Mol Cell Endocrinol 2001; 183 (Suppl 1): S77–S85.
Warner JP, Barron LH, Goudie D et al: A general method for the detection of large CAG repeat expansions by fluorescent PCR. J Med Genet 1996; 33: 1022–1026.
Coolbaugh-Murphy MI, Xu J, Ramagli LS, Brown BW, Siciliano MJ : Microsatellite instability (MSI) increases with age in normal somatic cells. Mech Ageing Dev 2005; 126: 1051–1059.
Pavlakis SG, Phillips PC, DiMauro S, De Vivo DC, Rowland LP : Mitochondrial myopathy, encephalopathy, lactic acidosis, and strokelike episodes: a distinctive clinical syndrome. Ann Neurol 1984; 16: 481–488.
Montagna P, Gallassi R, Medori R et al: MELAS syndrome: characteristic migrainous and epileptic features and maternal transmission. Neurology 1988; 38: 751–754.
Auvinen S, Suominen T, Hannonen P, Bachinski LL, Krahe R, Udd B : Myotonic dystrophy type 2 found in two of sixty-three persons diagnosed as having fibromyalgia. Arthritis Rheum 2008; 58: 3627–3631.
Suominen T, Schoser B, Raheem O et al: High frequency of co-segregating CLCN1 mutations among myotonic dystrophy type 2 patients from Finland and Germany. J Neurol 2008; 255: 1731–1736.
Arsenault ME, Prevost C, Lescault A, Laberge C, Puymirat J, Mathieu J : Clinical characteristics of myotonic dystrophy type 1 patients with small CTG expansions. Neurology 2006; 66: 1248–1250.
Schoser BG, Ricker K, Schneider-Gold C et al: Sudden cardiac death in myotonic dystrophy type 2. Neurology 2004; 63: 2402–2404.
Udd B, Vihola A, Sarparanta J, Richard I, Hackman P : Titinopathies and extension of the M-line mutation phenotype beyond distal myopathy and LGMD2J. Neurology 2005; 64: 636–642.
Acknowledgements
We acknowledge Tamer Ahmed for his technical assistance with DM2 and DM1 mutation analysis. We also acknowledge Merja Soininen and Satu Luhtasela for their assistance with sample preparations and logistics. TS was funded by the Finnish Cultural Foundation, Pirkanmaa Regional fund and the Pirkanmaa Hospital District research fund. CA was supported by the Telethon Genetic Biobank project GTB07001, the Eurobiobank project no. QLRI-CT-2002-02769 and the Cariparo grant. RK was supported by grants from the NIH (AR48171), the Muscular Dystrophy Association, USA and by the Kleberg Foundation. BU was supported by grants from the Vaasa Central Hospital district and Pirkanmaa Hospital district medical research funds and the Liv och Hälsa medical research foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Suominen, T., Bachinski, L., Auvinen, S. et al. Population frequency of myotonic dystrophy: higher than expected frequency of myotonic dystrophy type 2 (DM2) mutation in Finland. Eur J Hum Genet 19, 776–782 (2011). https://doi.org/10.1038/ejhg.2011.23
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2011.23
Keywords
This article is cited by
-
A retrospective study of accuracy and usefulness of electrophysiological exercise tests
Journal of Neurology (2024)
-
Multi-level profiling unravels mitochondrial dysfunction in myotonic dystrophy type 2
Acta Neuropathologica (2024)
-
A diagnostic support system based on pain drawings: binary and k-disease classification of EDS, GBS, FSHD, PROMM, and a control group with Pain2D
Orphanet Journal of Rare Diseases (2023)
-
Update on Therapy for Myotonic Dystrophy Type 1
Current Treatment Options in Neurology (2023)
-
Clinical score for early diagnosis of myotonic dystrophy type 2
Neurological Sciences (2023)